UCSF-PDGM | The University of California San Francisco Preoperative Diffuse Glioma MRI
DOI: 10.7937/tcia.bdgf-8v37 | Data Citation Required | 20k Views | 26 Citations | Image Collection
| Location | Species | Subjects | Data Types | Cancer Types | Size | Status | Updated | |
|---|---|---|---|---|---|---|---|---|
| Brain | Human | 495 | MR, Measurement, Other, Demographic, Follow-Up, Diagnosis, Molecular Test, Classification | Diffuse Glioma | Clinical, Genomics, Image Analyses, Software/Source Code | Public, Complete | 2023/04/07 |
Summary
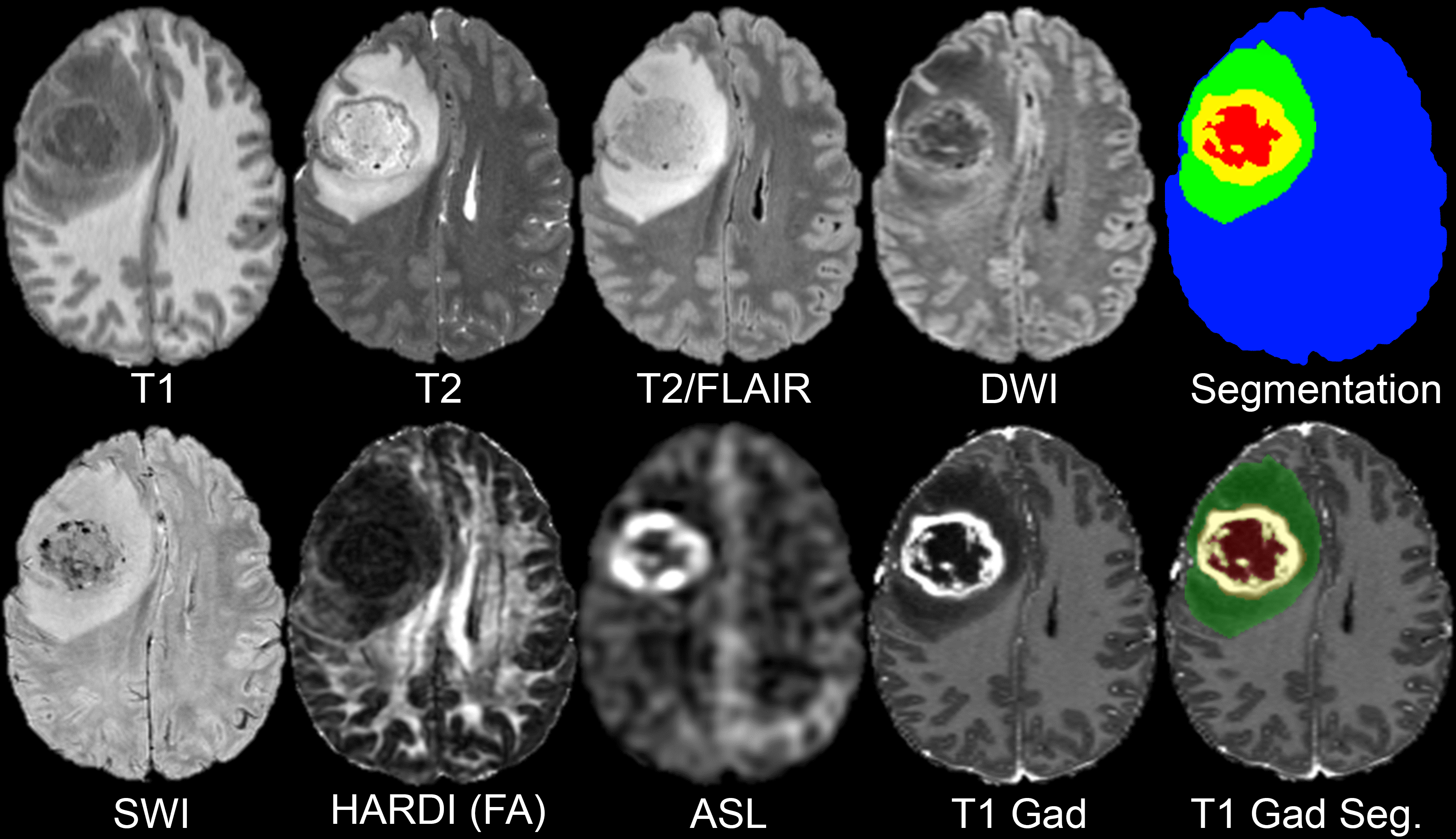
MRI-based artificial intelligence (AI) research on patients with brain gliomas has been rapidly increasing in popularity in recent years in part due to a growing number of publicly available MRI datasets. Notable examples include The Cancer Genome Atlas Glioblastoma dataset (TCGA-GBM) consisting of 262 subjects and the International Brain Tumor Segmentation (BraTS) challenge dataset consisting of 542 subjects (including 243 preoperative cases from TCGA-GBM). The public availability of these glioma MRI datasets has fostered the growth of numerous emerging AI techniques including automated tumor segmentation, radiogenomics, and MRI-based survival prediction. Despite these advances, existing publicly available glioma MRI datasets have been largely limited to only 4 MRI contrasts (T2, T2/FLAIR, and T1 pre- and post-contrast) and imaging protocols vary significantly in terms of magnetic field strength and acquisition parameters. Here we present the University of California San Francisco Preoperative Diffuse Glioma MRI (UCSF-PDGM) dataset. The UCSF-PDGM dataset includes 501 subjects with histopathologically-proven diffuse gliomas who were imaged with a standardized 3 Tesla preoperative brain tumor MRI protocol featuring predominantly 3D imaging, as well as advanced diffusion and perfusion imaging techniques. The dataset also includes isocitrate dehydrogenase (IDH) mutation status for all cases and O[6]-methylguanine-DNA methyltransferase (MGMT) promotor methylation status for World Health Organization (WHO) grade III and IV gliomas. The UCSF-PDGM has been made publicly available in the hopes that researchers around the world will use these data to continue to push the boundaries of AI applications for diffuse gliomas. Data collection was performed in accordance with relevant guidelines and regulations and was approved by the University of California San Francisco institutional review board with a waiver for consent. The dataset population consisted of 501* adult patients with histopathologically confirmed grade II-IV diffuse gliomas who underwent preoperative MRI, initial tumor resection, and tumor genetic testing at a single medical center between 2015 and 2021. Patients with any prior history of brain tumor treatment were excluded; however, history of tumor biopsy was not considered an exclusion criterion. All subjects’ tumors were tested for IDH mutations by genetic sequencing of tissue acquired during biopsy or resection. All grade III and IV tumors were tested for MGMT methylation status using a methylation sensitive quantitative PCR assay. The 501* cases included in the UCSF-PDGM include 55 (11%) grade II, 42 (9%) grade III, and 403 (80%) grade IV tumors. There was a male predominance for all tumor grades (56%, 60%, and 60%, respectively for grades II-IV). IDH mutations were identified in a majority of grade II (83%) and grade III (67%) tumors and a small minority of grade IV tumors (8%). MGMT promoter hypermethylation was detected in 63% of grade IV gliomas and was not tested for in a majority of lower grade gliomas. 1p/19q codeletion was detected in 20% of grade II tumors and a small minority of grade III (5%) and IV (<1%) tumors. Tabulated details and glossary are available in the Data Access and Detailed Description tabs below. All preoperative MRI was performed on a 3.0 tesla scanner (Discovery 750, GE Healthcare, Waukesha, Wisconsin, USA) and a dedicated 8-channel head coil (Invivo, Gainesville, Florida, USA). The imaging protocol included 3D T2-weighted, T2/FLAIR-weighted, susceptibility-weighted (SWI), diffusion-weighted (DWI), pre- and post-contrast T1-weighted images, 3D arterial spin labeling (ASL) perfusion images, and 2D 55-direction high angular resolution diffusion imaging (HARDI). Over the study period, two gadolinium-based contrast agents were used: gadobutrol (Gadovist, Bayer, LOC) at a dose of 0.1 mL/kg and gadoterate (Dotarem, Guerbet, Aulnay-sous-Bois, France) at a dose of 0.2 mL/kg. HARDI data were eddy current corrected and processed using the Eddy and DTIFIT modules from FSL 6.0.2 yielding isotropic diffusion weighted images (DWI) and several quantitative diffusivity maps: mean diffusivity (MD), axial diffusivity (AD), radial diffusivity (RD), and fractional anisotropy (FA). Eddy correction was performed with outlier replacement on and topup correction off. DTIFIT was performed with simple least squares regression. Each image contrast was registered and resampled to the 3D space defined by the T2/FLAIR image (1 mm isotropic resolution) using automated non-linear registration (Advanced Normalization Tools). Resampled co-registered data were then skull stripped using a previously described and publicly available deep-learning algorithm: https://www.github.com/ecalabr/brain_mask/. Multicompartment tumor segmentation of study data was undertaken as part of the 2021 BraTS challenge. Briefly, image data first underwent automated segmentation using an ensemble model consisting of prior BraTS challenge winning segmentation algorithms. Images were then manually corrected by trained radiologists and approved by 2 expert reviewers. Segmentation included three major tumor compartments: enhancing tumor, non-enhancing/necrotic tumor, and surrounding FLAIR abnormality (sometimes referred to as edema). The UCSF-PDGM adds to on an existing body of publicly available diffuse glioma MRI datasets that are commonly used in AI research applications. As MRI-based AI research applications continue to grow, new data are needed to foster development of new techniques and increase the generalizability of existing algorithms. The UCSF-PDGM not only significantly increases the total number of publicly available diffuse glioma MRI cases, but also provides a unique contribution in terms of MRI technique. The inclusion of 3D sequences and advanced MRI techniques like ASL and HARDI provides a new opportunity for researchers to explore the potential utility of cutting-edge clinical diagnostics for AI applications. In addition, these advanced imaging techniques may prove useful for radiogenomic studies focused on identification of IDH mutations or MGMT promoter methylation. The UCSF-PDGM dataset, particularly when combined with existing publicly available datasets, has the potential to fuel the next phase of radiologic AI research on diffuse gliomas. However, the UCSF-PDGM dataset’s potential will only be realized if the radiology AI research community takes advantage of this new data resource. We hope that this dataset sparks inspiration in the next generation of AI researchers, and we look forward to the new techniques and discoveries that the UCSF-PDGM will generate.Introduction
Methods
Patient Population
Genetic Biomarker Testing
Study participant demographic data
Image Acquisition
Image Pre-Processing
Tumor Segmentation
Data Access
Version 4: Updated 2023/04/07
Added bvec and bval to data access table. Added metadata glossary from Detailed description to Data Access table
| Title | Data Type | Format | Access Points | Subjects | License | Metadata | |||
|---|---|---|---|---|---|---|---|---|---|
| Images and Annotations | MR | NIFTI | Download requires IBM-Aspera-Connect plugin |
495 | 501 | 11,523 | CC BY 4.0 | — | |
| Clinical data | Demographic, Molecular Test, Diagnosis, Classification, Other | CSV | CC BY 4.0 | — | |||||
| Clinical metadata glossary | Other, Demographic, Follow-Up, Diagnosis, Molecular Test | CSV | CC BY 4.0 | — | |||||
| bval file | Measurement | BVAL and ZIP | CC BY 4.0 | — | |||||
| bvec file | Measurement | BVEC and ZIP | CC BY 4.0 | — |
Citations & Data Usage Policy
Data Citation Required: Users must abide by the TCIA Data Usage Policy and Restrictions. Attribution must include the following citation, including the Digital Object Identifier:
Data Citation |
|
|
Calabrese, E., Villanueva-Meyer, J., Rudie, J., Rauschecker, A., Baid, U., Bakas, S., Cha, S., Mongan, J., & Hess, C. (2022). The University of California San Francisco Preoperative Diffuse Glioma MRI (UCSF-PDGM) (Version 4) [Dataset]. The Cancer Imaging Archive. DOI: 10.7937/tcia.bdgf-8v37 |
Detailed Description
*Note: it was discovered that some ID are followup imaging of others, therefore the true number of patients in this Collection is 495 not 501, the pixels have not been changed:
UCSF-PDGM-0433 was imaged 7 days later as UCSF-PDGM-0315 (version 3 files reflect this change by renaming UCSF-PDGM-0315 to UCSF-PDGM-0433_FU007d)
UCSF-PDGM-0431 was imaged 1 days later as UCSF-PDGM-0278 (version 3 files reflect this change by renaming UCSF-PDGM-0278 to UCSF-PDGM-0431_FU001d)
UCSF-PDGM-0396 was imaged 175 days later as UCSF-PDGM-0175 (version 3 files reflect this change by renaming UCSF-PDGM-0175 to UCSF-PDGM-0396_FU175d)
UCSF-PDGM-0429 was imaged 3 days later as UCSF-PDGM-0138 (version 3 files reflect this change by renaming UCSF-PDGM-0138 to UCSF-PDGM-0429_FU003d)
UCSF-PDGM-0409 was imaged 1 days later as UCSF-PDGM-0181 (version 3 files reflect this change by renaming UCSF-PDGM-0181 to UCSF-PDGM-0409_FU001d)
UCSF-PDGM-0391 was imaged 16 days later as UCSF-PDGM-0289 (version 3 files reflect this change by renaming UCSF-PDGM-0289 UCSF-PDGM-0391_FU016d)
All image data have been “skull stripped”, deidentified, pre-processed per the methods section of our abstract, and converted to NIfTI format. We cannot provide original DICOM data, however, these pre-processed files have been prepared to facilitate the type of research that this dataset is intended for. Publicly available deep-learning algorithm for performing this process is here: https://www.github.com/ecalabr/brain_mask/.
Glossary of abbreviations: UCSF-PDGM-metadata_glossary.csv
| Term | Represents | Values |
|---|---|---|
| ID | DICOM (0010,0020) PatientID | |
| Sex | DICOM (0010,0040) Patient Sex | M,F |
| Age at MRI | Age in years at time of MR imaging | |
| WHO CNS Grade | Grade per the 2021 World Health Organization Classification of Tumors of the Central Nervous System (WHO CNS 2021) (https://doi.org/10.1093/neuonc/noab106 ) | 2,3,4 |
| Final pathologic diagnosis (WHO 2021) | Final (integrated) pathologic diagnosis per the 2021 World Health Organization Classification of Tumors of the Central Nervous System (WHO CNS 2021) ( https://doi.org/10.1093/neuonc/noab106 ) |
|
| MGMT status | O6-methylguanine-DNA methyltransferase status – clinical interpretation of the MGMT index described below. | negative, positive, indeterminate |
| MGMT index | O6-methylguanine-DNA methyltransferase methylation index (in house method developed by UCSF clinical labs, https://genomics.ucsf.edu/content/mgmt-promoter-methylation-assay ) where numeric values 0-17 indicate the number of promoter methylation sites. | 0-17, blank |
| 1p/19q | presence of codeletion of 1p and 19q genes, assayed by fluorescent in-situ hybridization | intact, co-deletion, relative co-deletion, unknown |
| IDH | isocitrate dehydrogenase mutation subtype characterized with a capture-based targeted next-generation DNA sequencing panel (UCSF500) as described in (https://doi.org/10.1093/neuonc/now254 ) | |
| 1-dead 0-alive | Survival status of the patient at last clinical follow up. | |
| OS | Overall survival in days from initial diagnosis to last clinical follow up. | |
| EOR | Extent of resection determined by review of operative reports and immediate postoperative imaging | biopsy (only biopsy) Subtotal resection (STR) gross total resection (GTR) |
| Biopsy prior to imaging | Was a burr-hole biopsy performed prior to imaging | yes, no, blank |
Note that The L1/L2/L3 files the eigenvalues. These are direct outputs from FSL DTIFIT as described here: https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FDT/UserGuide#DTIFIT . We elected not to include the tensor files since they are quite large and are straightforward to derive from the provided 4D DWI data using FSL DTIFIT. The link provided above also includes instructions for how to do this in case the user is not familiar.
Acknowledgements
We would like to acknowledge the individuals and institutions that have provided data for this collection:
Research was supported by the National Institutes of Health Ruth L. Kirschstein Institutional National Research Service Award under award number T32EB001631 and by the RSNA Research & Education Foundation under grant number RR2011. The content is solely the responsibility of the authors and does not necessarily represent the official views of the RSNA R&E Foundation.
Related Publications
Publications by the Dataset Authors
The authors recommended the following as the best source of additional information about this dataset:
Publication Citation |
|
|
Evan Calabrese, Javier E. Villanueva-Meyer, Jeffrey D. Rudie, Andreas M. Rauschecker, Ujjwal Baid, Spyridon Bakas, Soonmee Cha, John T. Mongan, Christopher P. Hess. (2022) The UCSF Preoperative Diffuse Glioma MRI (UCSF-PDGM) Dataset. Radiology: Artificial Intelligence. DOI: https://doi.org/10.1148/ryai.220058 |
No other publications were recommended by dataset authors.
Research Community Publications
TCIA maintains a list of publications that leveraged this dataset. If you have a manuscript you’d like to add please contact TCIA’s Helpdesk.
Previous Versions
Version 3: Updated 2023/01/11
*Note: it was discovered that some ID are followup imaging of others, therefore the true number of patients in this Collection is 495, the pixels have not been changed:
UCSF-PDGM-0433 was imaged 7 days later as UCSF-PDGM-0315 (version 3 files reflect this change by renaming UCSF-PDGM-0315 to UCSF-PDGM-0433_FU007d)
UCSF-PDGM-0431 was imaged 1 day later as UCSF-PDGM-0278 (version 3 files reflect this change by renaming UCSF-PDGM-0278 to UCSF-PDGM-0431_FU001d)
UCSF-PDGM-0396 was imaged 175 days later as UCSF-PDGM-0175 (version 3 files reflect this change by renaming UCSF-PDGM-0175 to UCSF-PDGM-0396_FU175d)
UCSF-PDGM-0429 was imaged 3 days later as UCSF-PDGM-0138 (version 3 files reflect this change by renaming UCSF-PDGM-0138 to UCSF-PDGM-0429_FU003d)
UCSF-PDGM-0409 was imaged 1 day later as UCSF-PDGM-0181 (version 3 files reflect this change by renaming UCSF-PDGM-0181 to UCSF-PDGM-0409_FU001d)
UCSF-PDGM-0391 was imaged 16 days later as UCSF-PDGM-0289 (version 3 files reflect this change by renaming UCSF-PDGM-0289 UCSF-PDGM-0391_FU016d)
| Title | Data Type | Format | Access Points | License | Metadata | ||||
|---|---|---|---|---|---|---|---|---|---|
| Images and Annotations | NIFTI | Download requires IBM-Aspera-Connect plugin |
CC BY 4.0 | — | |||||
| Clinical data | CSV | CC BY 4.0 | — |
Version 2: Updated 2022/11/30
Changes to this version:
- Fixes to integer rounding errors in *tumor_segmentation.nii.gz files by the Collection’s investigators.
- TCIA users have asked for a mapping to the 2021 BraTS data to prevent any data leak when both datasets are used. Updated metadata spreadsheet attached which includes the BraTS IDs for all relevant cases. This data was confirmed by BraTS Challenge organizers.
- Correction of name suffix for bias corrected T1 postcontrast images from “T1gad_bias” to “T1c_bias” for consistency.
| Title | Data Type | Format | Access Points | License | Metadata | ||||
|---|---|---|---|---|---|---|---|---|---|
| Images and Annotations | NIFTI | Download requires IBM-Aspera-Connect plugin |
CC BY 4.0 | — | |||||
| Clinical data | CSV | CC BY 4.0 | — |
Version 1: Updated 2022/09/26
| Title | Data Type | Format | Access Points | License | Metadata | ||||
|---|---|---|---|---|---|---|---|---|---|
| Images and Annotations | NIFTI and ZIP | — | |||||||
| Clinical data | CSV | CC BY 4.0 | — |
