PleThora | Thoracic Volume and Pleural Effusion Segmentations in Diseased Lungs for Benchmarking Chest CT Processing Pipelines
DOI: 10.7937/tcia.2020.6c7y-gq39 | Data Citation Required | 1.6k Views | 7 Citations | Analysis Result
| Location | Subjects | Size | Updated | ||
|---|---|---|---|---|---|
| Lung | Lung | 402 | 2020/07/28 |
Summary
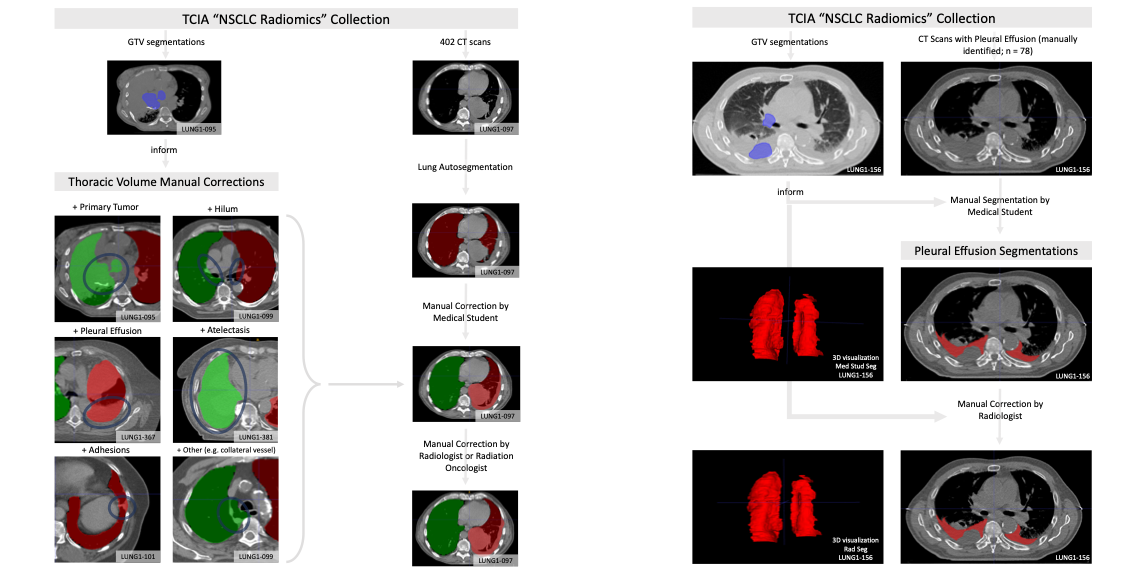
Automated or semi-automated algorithms intended for chest CT analyses typically require the creation of a 3D map of the thoracic volume as their initial step. Identifying this anatomic region precedes fundamental tasks such as lung structure segmentation, lesion detection, and radiomics feature extraction in analysis pipelines. However, automatic approaches to segment the thoracic volume maps struggle to perform consistently in subjects with diseased lungs – yet this is exactly the circumstance for which pipeline analyses would be most useful. To address this need, we have created PleThora, a dataset of pleural effusion and thoracic cavity segmentations in subjects with diseased lungs. PleThora consists of left and right thoracic cavity segmentations delineated on 402 CT scans from The Cancer Imaging Archive NSCLC-Radiomics collection as well as separate segmentations labeling pleural effusions alone. Thoracic cavity segmentations include lung parenchyma, tumor, atelectasis, adhesions, and effusion. PleThora is a tool for medical image preprocessing and segmentation researchers to build and compare approaches for region-of-interest identification and analysis. The thoracic cavity segmentations were generated automatically by a U-Net based algorithm trained on chest CTs without cancer, manually corrected by a medical student, and revised by a radiation oncologist or a radiologist. Pleural effusion segmentations were manually delineated by a medical student and revised by a radiologist. Expert GTV segmentations already provided by the NSCLC-Radiomics collection helped inform our segmentations, and areas of the effusion that overlap with GTVs are not included. Researchers interested in discriminating between GTV and effusion using imaging biomarker inputs may find our pleural effusion segmentations useful, especially when paired with the GTV segmentations provided in the NSCLC-Radiomics collection. Tabular data are also provided, including GTV, thorax, and effusion volumes (in cm3), tumor location, and image metadata. Additionally, we standardized a train/test split for training deep learning algorithms with the thoracic cavity segmentations. Note: These segmentations use the RPI orientation, but the original DICOM files are oriented using the RAI convention. As a result, some tools such as ITK-SNAP will not render the segmentations in the correct orientation when visualized. The authors of these data suggest using software like Mango (http://ric.uthscsa.edu/mango/) or to convert to DICOM files to NIfTI with software like dcm2niix (https://github.com/rordenlab/dcm2niix) to address this issue.
Data Access
Version 3: Updated 2020/07/28
Version 3 changes:
2D U-Net
- Incorrectly reported the 2D U-Net achieved segmentations with Dice similarity coefficients of 0.90 and 0.94 for left and right lungs.
- The performances should be 0.94 and 0.94.
3D U-Net
- Incorrectly reported the 3D U-Net achieved segmentations with Dice similarity coefficients of 0.82 and 0.94 for left and right lungs.
- The performances should be 0.95 and 0.96.
Data Dictionary
- Added Auto-MS Thorax DSC description.
| Title | Data Type | Format | Access Points | Subjects | License | Metadata | |||
|---|---|---|---|---|---|---|---|---|---|
| Thoracic Segmentations | Segmentation | NIFTI and ZIP | 402 | 402 | CC BY 3.0 | — | |||
| Pleural Effusion Segmentations | Segmentation | NIFTI and ZIP | 78 | 172 | CC BY 3.0 | — | |||
| Segmentation Features and Image Metadata | Radiomic Feature, Measurement | CSV | CC BY 3.0 | — | |||||
| Baseline UNet 2D Summary | Other | CC BY 3.0 | — | ||||||
| Baseline UNet 3D Summary | Other | CC BY 3.0 | — | ||||||
| Data Dictionary | Other | DOCX | CC BY 3.0 | — |
Collections Used In This Analysis Result
| Title | Data Type | Format | Access Points | Subjects | License | Metadata | |||
|---|---|---|---|---|---|---|---|---|---|
| Corresponding Original CT Images from NSCLC-Radiomics | CT | DICOM | Requires NBIA Data Retriever |
402 | 402 | 402 | 48,568 | CC BY 3.0 | View |
Citations & Data Usage Policy
Data Citation Required: Users must abide by the TCIA Data Usage Policy and Restrictions. Attribution must include the following citation, including the Digital Object Identifier:
Data Citation |
|
|
Kiser, K.J., Ahmed, S., Stieb, S.M., Mohamed, A.S.R., Elhalawani, H., Park, P.Y.S., Doyle, N.S., Wang, B.J., Barman, A., Fuller, C.D., Giancardo, L. (2020). Data from the Thoracic Volume and Pleural Effusion Segmentations in Diseased Lungs for Benchmarking Chest CT Processing Pipelines (PleThora) [Data set]. The Cancer Imaging Archive. https://doi.org/10.7937/tcia.2020.6c7y-gq39 . |
Data Citation |
|
|
Aerts, H. J. W. L., Wee, L., Rios Velazquez, E., Leijenaar, R. T. H., Parmar, C., Grossmann, P., Carvalho, S., Bussink, J., Monshouwer, R., Haibe-Kains, B., Rietveld, D., Hoebers, F., Rietbergen, M. M., Leemans, C. R., Dekker, A., Quackenbush, J., Gillies, R. J., & Lambin, P. (2019). Data From NSCLC-Radiomics (Version 4) [Data set]. The Cancer Imaging Archive. https://doi.org/10.7937/K9/TCIA.2015.PF0M9REI |
Acknowledgement |
|
|
Swiss Cancer League (BIL KLS-4300-08-2017). |
Acknowledgement |
|
|
Learning Healthcare Award funded by the UTHealth Center for Clinical and Translational Science (CCTS). |
Acknowledgement |
|
|
NIH grant UL1TR003167. |
Acknowledgement |
|
|
National Institutes of Health (NIH) National Institute for Dental and Craniofacial Research Establishing Outcome Measures Award (1R01DE025248/R56DE025248) and an Academic Industrial Partnership Grant (R01DE028290) |
Acknowledgement |
|
|
National Cancer Institute (NCI) Early Phase Clinical Trials in Imaging and Image-Guided Interventions Program (1R01CA218148) |
Acknowledgement |
|
|
NIH/NCI Cancer Center Support Grant (CCSG) Pilot Research Program Award from the UT MD Anderson CCSG Radiation Oncology and Cancer Imaging Program (P30CA016672) |
Acknowledgement |
|
|
NIH/NCI Head and Neck Specialized Programs of Research Excellence (SPORE) Developmental Research Program Award (P50 CA097007) |
Acknowledgement |
|
|
National Science Foundation (NSF), Division of Mathematical Sciences, Joint NIH/NSF Initiative on Quantitative Approaches to Biomedical Big Data (QuBBD) Grant (NSF 1557679) |
Acknowledgement |
|
|
NSF Division of Civil, Mechanical, and Manufacturing Innovation (CMMI) standard grant (NSF 1933369) a National Institute of Biomedical Imaging and Bioengineering (NIBIB) Research Education Programs for Residents and Clinical Fellows Grant (R25EB025787-01) |
Acknowledgement |
|
|
NIH Big Data to Knowledge (BD2K) Program of the NCI Early Stage Development of Technologies in Biomedical Computing, Informatics, and Big Data Science Award (1R01CA214825). |
Acknowledgement |
|
|
Direct infrastructure support was provided by the multidisciplinary Stiefel Oropharyngeal Research Fund of the University of Texas MD Anderson Cancer Center Charles and Daneen Stiefel Center for Head and Neck Cancer and the Cancer Center Support Grant (P30CA016672) and the MD Anderson Program in Image-guided Cancer Therapy. |
Acknowledgement |
|
|
Direct industry grant support, honoraria, and travel funding from Elekta AB |
Detailed Description
All NIfTI files have been compressed for convenience (.nii.gz)
Note: These segmentations use the RPI orientation, but the original DICOM files are oriented using the RAI convention. As a result, some tools such as ITK-SNAP will not render the segmentations in the correct orientation when visualized. The authors of these data suggest using software like Mango (http://ric.uthscsa.edu/mango/) or to convert to DICOM files to NIfTI with software like dcm2niix (https://github.com/rordenlab/dcm2niix) to address this issue.
Acknowledgements
We would like to acknowledge the individuals and institutions that have provided data for this collection:
- University of Texas M.D. Anderson Cancer Center, Houston, TX, USA – Special thanks to Kendall Kiser, MS Biomedical Informatics, from the Department of Radiation Oncology.
- The University of Texas Health Science Center School of Biomedical Informatics, Houston, TX, USA
- John P. and Kathrine G. McGovern Medical School, Houston, TX. Department of Diagnostic and Interventional Imaging.
Related Publications
Publications by the Dataset Authors
The authors recommended the following as the best source of additional information about this dataset:
Publication Citation |
|
|
Kiser, K.J., Barman, A., Stieb, S., Fuller, C.D., Giancardo, L., 2021. Novel Autosegmentation Spatial Similarity Metrics Capture the Time Required to Correct Segmentations Better Than Traditional Metrics in a Thoracic Cavity Segmentation Workflow. J Digit Imaging. https://doi.org/10.1007/s10278-021-00460-3 |
Research Community Publications
TCIA maintains a list of publications that leveraged this dataset. If you have a manuscript you’d like to add please contact TCIA’s Helpdesk.
Previous Versions
Version 2: Updated 2020/06/26
Version 2 changes:
- The dataset is now named “PleThora” for “Pleural effusion and thoracic cavity segmentations in diseased lungs.”
- All NIfTI files have been compressed for convenience (.nii à .nii.gz)
- All thoracic cavity primary reviewer segmentations have been renamed from “lungMask_edit.nii” to “[CaseID]_thor_cav_primary_reviewer.nii.gz” to more specifically identify each file’s contents and avoid confusion.
- Eighty-six thoracic cavity secondary reviewer segmentations have been added. These are named “[CaseID]_thor_cav_secondary_reviewer.nii.gz.”
- Interobserver variability analysis between primary and secondary reviewer thoracic cavity segmentations revealed four cases in which interobserver agreement was anomalously lower than all other cases. These cases were manually re-reviewed by another physician. In three cases (LUNG1-026, LUNG1-157, and LUNG1-354) it was deemed that the secondary reviewer’s segmentation excluded structures that should have been included. These were corrected. In one case (LUNG-088) it was determined that the primary reviewer segmentation included a large (400 cm3) nodal conglomerate. Our original thoracic cavity segmentation definition did not intend to include nodal conglomerates, so for consistency’s sake we corrected the primary reviewer segmentation accordingly. However, the segmentation with the nodal conglomerate is still valuable, so we provide it as well and name it “LUNG1-088_thor_cav_primary_reviewer_with_nodal_conglomerate.nii”
- We manually reviewed the pleural effusion segmentations of the primary physician reviewer and determined that in many cases the reviewer had not been sufficiently careful. Therefore, all 78 primary reviewer segmentations were re-reviewed by another physician and corrected as necessary. They are now re-submitted as “[CaseID]_effusion_first_reviewer.nii.gz”
- Seventy-eight pleural effusion secondary reviewer segmentations have been added. These are named “[CaseID]_effusion_second_reviewer.nii.gz.”
- Fifteen pleural effusion tertiary reviewer segmentations have been added. These are named “[CaseID]_effusion_third_reviewer.nii.gz.”
- We add two documents that describe baseline performances for 2D and 3D U-Net segmentation algorithms and define a reproducible train/test split.
- Data Dictionary: we provide a data dictionary to describe the meanings of column names in the “Thorax and Pleural Effusion Segmentation Metadata” spreadsheet.
