AAPM-RT-MAC | AAPM RT-MAC Grand Challenge 2019
DOI: 10.7937/tcia.2019.bcfjqfqb | Data Citation Required | 1.2k Views | 8 Citations | Image Collection
| Location | Species | Subjects | Data Types | Cancer Types | Size | Status | Updated |
|---|---|---|---|---|---|---|---|
| Head-Neck | Human | 55 | MR, RTSTRUCT | Head and Neck Cancer | Limited, Complete | 2020/07/20 |
Summary
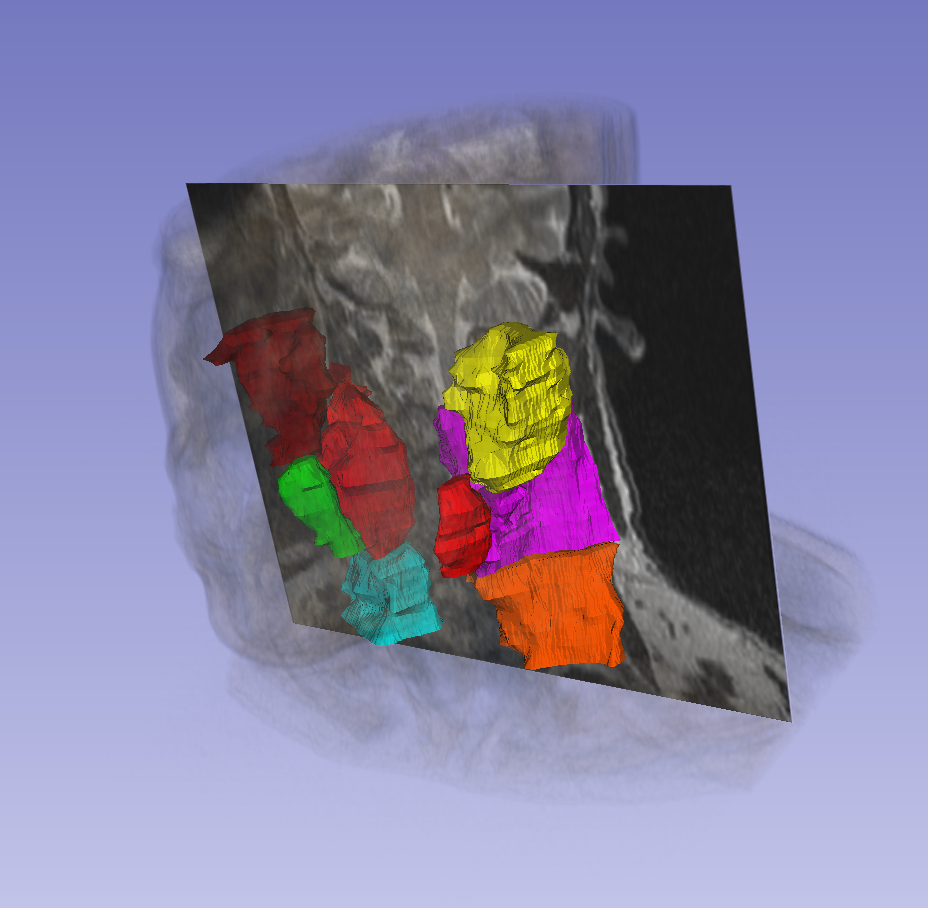
This data set was provided in association with a challenge competition and related conference session conducted at the AAPM 2019 Annual Meeting. MRI is popular in radiation oncology because of its excellent imaging quality of soft tissue and tumor. With the advent of MR-Linac and MR-guided radiation therapy, there is a trend toward a MR-based radiation treatment planning. Contouring is an important task in modern radiation treatment planning and frequently introduces uncertainties in radiation therapy due to observer variabilities. Auto-segmentation has been demonstrated as an effective approach to reduce this uncertainty. The overall objective of this grand challenge is to provide a platform for comparison of various auto-segmentation algorithms when they are used to delineate organs at risk (OARs) or tumors from MR images for head and neck patients for radiation treatment planning. The results will provide an indication of the performances achieved by various auto-segmentation algorithms and can be used to guide the selection of these algorithms for clinic use if desirable. The data for this challenge contains a total of 55 MRI cases, each from a single examination from a distinct patient, with each case consisting of a T2-weighted MRI images in DICOM format. The MRI scanning protocol was designed for radiation treatment simulation. Thirty-one of these will be provided as training cases, with the parotid glands, submandibular glands, level 2 and level 3 lymph nodes contoured. The images and contours were acquired from MD Anderson Cancer Center. More details on accessing the various challenge subsets (training, off-site test, and live test) can be found on the Detailed Description tab below.
Data Access
Some data in this collection contains images that could potentially be used to reconstruct a human face. To safeguard the privacy of participants, users must sign and submit a TCIA Restricted License Agreement to help@cancerimagingarchive.net before accessing the data.
Version 3: Updated 2020/07/20
RTSTRUCTS for test and live data, previously embargoed, are now publicly visible. 12 RTSTRUCTS Test subjects, 12 RTSTRUCTS Live subjects.
| Title | Data Type | Format | Access Points | Subjects | License | Metadata | |||
|---|---|---|---|---|---|---|---|---|---|
| Images and Radiation Therapy Structures | MR, RTSTRUCT | DICOM | 55 | 55 | 110 | 6,655 | NIH Controlled Data Access Policy | View |
Citations & Data Usage Policy
Data Citation Required: Users must abide by the TCIA Data Usage Policy and Restrictions. Attribution must include the following citation, including the Digital Object Identifier:
Data Citation |
|
|
Cardenas, C., Mohamed, A., Sharp, G., Gooding, M., Veeraraghavan, H., & Yang, J. (2019). Data from AAPM RT-MAC Grand Challenge 2019. The Cancer Imaging Archive. https://doi.org/10.7937/tcia.2019.bcfjqfqb. |
Detailed Description
Supporting Documentation and Metadata
Some information from the 2019 challenge site https://www.aapm.org/GrandChallenge/RT-MAC/ is included below.
Data description
T2 MR Images of 55 patients have been collected for this challenge. Data were acquired from MD Anderson Cancer Center. All images were scanned with a protocol specifically designed for head-and-neck radiation treatment simulation. Datasets were divided into three groups:
- 31 training datasets
- 12 off-site test datasets
- 12 live test datasets
Data will be provided in DICOM (both MR and RTSTRUCT), as commonly used in most commercial treatment planning systems.
Contouring Guidelines
The manual contours were drawn by a radiation oncologist at MD Anderson Cancer Center. The structures were contoured following the DAHANCA, EORTC, GORTEC, HKNPCSG, NCIC CTG, NCRI, NRG Oncology and RROG consensus guidelines reported in Brouwer et.al Radiotherapy and Oncol. 2015. Details of contouring guidelines can be found in “Learn the Details” on the challenge website. The following structures are included in this challenge:
- Parotid glands
- Submandibular glands
- Lymph nodes level II
- Lymph nodes level III
Training Data
Each training dataset includes a set of DICOM MR image files and one DICOM RTSTRUCT file. Each training dataset is labeled as RTMAC-TRAIN-xxx, where xxx identifies the dataset ID.
Training data are available here. (Requires NBIA Data Retriever.)
Off-site test data
Each off-site test dataset includes a set of DICOM MR image files and is labeled as RTMAC-TEST-xxx, where xxx identifies the dataset ID.
Off-site test data are available here. (Requires NBIA Data Retriever.)
RTSTRUCT data associated with Off-site test data were made available 7/20/2020.
Live test data
Each Live test dataset includes a set of DICOM MR image files and is labeled as RTMAC-LIVE-xxx, where xxx identifies the dataset ID.
Live test data are available here. (Requires NBIA Data Retriever.)
RTSTRUCT data associated with Live test data were made available 7/20/2020.
Acknowledgements
We would like to acknowledge the individuals and institutions that have provided data for this collection:
- The University of Texas MD Anderson Cancer Center Houston, Texas, USA
Related Publications
Publications by the Dataset Authors
The authors recommended the following as the best source of additional information about this dataset:
Publication Citation |
|
|
Cardenas, C.E., Mohamed, A.S.R., Yang J., Gooding, M., Veeraraghavan, H., Kalpathy-Cramer J., Ng S.P., Ding Y., Wang J., Lai S.Y., Fuller C.D., & Sharp, G. (2020) Head and neck cancer patient images for determining auto-segmentation accuracy in T2-weighted magnetic resonance imaging through expert manual segmentations. Med. Phys, 47(5):2317-2322. DOI: https://doi.org/10.1002/mp.13942.
|
No other publications were recommended by dataset authors.
Research Community Publications
TCIA maintains a list of publications that leveraged this dataset. If you have a manuscript you’d like to add please contact TCIA’s Helpdesk.
Previous Versions
Version 2: Updated 2019/07/15
Release of live data for AAPM challenge.
| Title | Data Type | Format | Access Points | License | Metadata | ||||
|---|---|---|---|---|---|---|---|---|---|
| Images MR | MR, RTSTRUCT | DICOM | Download requires NBIA Data Retriever |
55 | 55 | 86 | 6,631 | — |
Version 1: Updated 2019/05/23
| Title | Data Type | Format | Access Points | License | Metadata | ||||
|---|---|---|---|---|---|---|---|---|---|
| Images | MR, RTSTRUCT | DICOM | Download requires NBIA Data Retriever |
43 | 43 | 74 | 5,191 | — |
