ACRIN-6698 | ACRIN 6698/I-SPY2 Breast DWI
DOI: 10.7937/tcia.kk02-6d95 | Data Citation Required | 5.4k Views | 18 Citations | Image Collection
| Location | Species | Subjects | Data Types | Cancer Types | Size | Status | Updated | |
|---|---|---|---|---|---|---|---|---|
| Breast | Human | 385 | MR, SEG, Demographic, Molecular Test, Classification, Follow-Up, Other | Breast Cancer | Clinical, Image Analyses | Public, Complete | 2021/12/17 |
Summary
The American College of Radiology Imaging Network (ACRIN) trial 6698 (NCT01564368) was a multi-center study to evaluate the effectiveness of quantitative diffusion weighted imaging (DWI) for assessing breast cancer response to neoadjuvant chemotherapy (NAC). ACRIN 6698 was performed as a sub-study of the ongoing I-SPY 2 TRIAL (Investigation of Serial studies to Predict Your Therapeutic Response with Imaging And moLecular Analysis 2), an adaptive, multi-agent phase II trial designed to quickly identify new agents for breast cancer. Patients recruited for the I-SPY 2 TRIAL and enrolling at sites meeting DWI qualification requirements were eligible for the 6698 trial. 406 women with invasive breast cancer were prospectively enrolled to ACRIN 6698 at ten institutions between August 2012 to January 2015, and 272 were randomized to I‑SPY 2 experimental treatment or control arms. Patients underwent breast DWI using a 4-b value protocol, as well as standard T2-weighted and dynamic contrast enhanced (DCE) scans. MRI studies were conducted at 4 timepoints over the course of NAC: pre-treatment (T0), early-treatment after 3 cycles paclitaxel (T1), mid-treatment between paclitaxel and AC (T2) and post-treatment (T3). Of the 272 treated patients, 242 comprised the primary analysis cohort (30 were excluded for missing or non-evaluable DWI exams). This TCIA collection includes all MRI studies received by the UCSF image analysis lab, excluding studies with no analyzable acquisitions. A separate download option is provided to access only those studies in the primary analysis cohort. In addition, DWI test/retest data acquired at baseline or early-treatment in a subset of patients is included in the full collection and is available as a separate download option (N=89 subjects consented and imaged, 71 analyzable test/retest acquisition pairs). The ACRIN 6698 image data set is currently a unique collection for investigating the utility of DWI for monitoring of response to neoadjuvant breast cancer treatment. While many smaller and/or single-site DWI studies have been published, the multi-center and quality-control aspects of this data allow investigators true evaluation of analysis techniques in the clinical trial environment. The multi-b value protocol also allows evaluation of higher order diffusion models and evaluation at two different clinically relevant b values (600 and 800 s/mm2). Furthermore, the embedded test-retest arm of the study will allow evaluation of repeatability and reproducibility of new DWI metrics and analysis techniques. In addition to the original DWI data the collection includes derived ADC maps with manually delimited tumor segmentations from the primary study analysis (for all studies rated as analyzable in the QC evaluation), plus T2-weighted images, DCE images with derived enhancement maps, clinical data and outcome data (pathologic complete response [pCR] at surgery). Additional information about the trial is available in the Study Protocol and Case Report Forms.
Data Access
Version 1: Updated 2021/12/17
| Title | Data Type | Format | Access Points | Subjects | License | Metadata | |||
|---|---|---|---|---|---|---|---|---|---|
| Images and Segmentations | MR, SEG | DICOM | Download requires NBIA Data Retriever |
385 | 1,123 | 18,747 | 2,911,334 | CC BY 4.0 | View |
| ACRIN 6698 Primary Analysis Subgroup | MR, SEG | DICOM | 242 | 865 | 14,678 | 2,238,511 | CC BY 4.0 | View | |
| ACRIN 6698 Test-retest set | MR, SEG | DICOM | 71 | 71 | 1,573 | 231,844 | CC BY 4.0 | View | |
| BMMR2 Training set | MR, SEG | DICOM | 117 | 351 | 6,003 | 900,863 | CC BY 4.0 | View | |
| BMMR2 Testing set | MR, SEG | DICOM | 74 | 222 | 3,836 | 576,684 | CC BY 4.0 | View | |
| Full Collection Ancillary Patient Information file | Demographic, Molecular Test, Classification, Follow-Up, Other | XLSX | 385 | CC BY 4.0 | — |
Additional Resources for this Dataset
The NCI Cancer Research Data Commons (CRDC) provides access to additional data and a cloud-based data science infrastructure that connects data sets with analytics tools to allow users to share, integrate, analyze, and visualize cancer research data.
- Imaging Data Commons (IDC) (Imaging Data)
Citations & Data Usage Policy
Data Citation Required: Users must abide by the TCIA Data Usage Policy and Restrictions. Attribution must include the following citation, including the Digital Object Identifier:
Data Citation |
|
|
Newitt, D. C., Partridge, S. C., Zhang, Z., Gibbs, J., Chenevert, T., Rosen, M., Bolan, P., Marques, H., Romanoff, J., Cimino, L., Joe, B. N., Umphrey, H., Ojeda-Fournier, H., Dogan, B., Oh, K. Y., Abe, H., Drukteinis, J., Esserman, L. J., & Hylton, N. M. (2021). ACRIN 6698/I-SPY2 Breast DWI [Data set]. The Cancer Imaging Archive. https://doi.org/10.7937/tcia.kk02-6d95 |
Detailed Description
MRI Studies
Trial time points for MRI studies
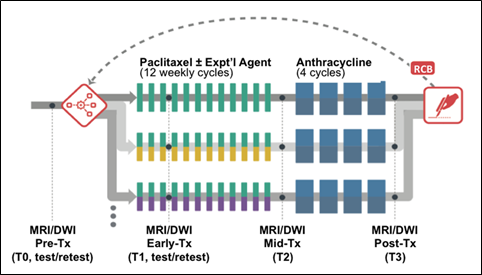
The primary aim of ACRIN 6698 was the evaluation of breast diffusion weighted imaging (DWI) for the prediction of response to neoadjuvant chemotherapy (NAC) for invasive breast cancer. For this purpose, serial MRI studies were acquired over the course of treatment. The study schema for the ACRIN 6698 Trial is shown in Figure 1
MRI studies were performed at up to four time points in the course of NAC:
- Pre-treatment (T0, optional test/retest visit)
Prior to randomization patients received an MRI study including T2weighted, 4 b‑value DWI, and dynamic contrast-enhanced (DCE) acquisitions.
For patients randomized onto a treatment arm of I‑SPY 2 subsequent MRI studies with the same required acquisitions were performed:
- Early-treatment (T1, optional test/retest visit)
After 3 weeks of regimen 1 treatment (Paclitaxel with or without an experimental agent). - Mid-treatment (T2)
Between Paclitaxel and Anthracycline (AC) treatment regimens - Post-treatment (T3)
Following 4 cycles AC and prior to surgery.
“Coffee-break” style test/retest studies were done on a subset of patients under a separate informed consent. These were by preference done at the baseline visit but were also allowed at visit T1. A given patient could only have the test/retest scans at a single visit, and the total number of test/retest cases at any given site was limited (maximum 14) to ensure a wide representation of different scanners, sites and field strengths in the test/retest cohort.
Figure 1. ACRIN 6698 study schema.
Imaging protocol and included original image series
MR imaging was performed on a 1.5 or 3.0 Tesla scanner using a dedicated breast radiofrequency coil. All studies for a given patient were required to be performed on the same scanner configuration (model, field strength and breast coil model). Subjects were imaged in the prone position. The image acquisition protocol included a localization scan and a minimum of three required acquisitions:
- T2weighted sequence
- DWI (b=0, 100, 600, 800 s/mm2, 3-direction)
- T1weighted DCE (80sec < phase duration < 100sec with at least 8 minutes continuous post-injection acquisition)
Protocol limits on scan parameters for these required acquisitions are given in an accompanying document ACRIN 6698 MRI Acquisition Parameters.
All required imaging was performed axially with full bilaterally coverage. For the TCIA collection all original images from these three acquisitions are included. Derived images submitted with the study including reformats, CAD software outputs and MIP images are not part of the collection. For the T2weighted acquisitions multiple series are included if they were generated as part of the standard reconstruction process on the scanner. For example, acquisitions utilizing Dixon techniques to create separate fat and water images will have multiple series in the collection. All images in the original DWI series are included, including directional diffusion images for cases where they were retained and submitted with the study. For some General Electric Medical Systems (GEMS) scanners employed in the trial the acquisition software allowed only 2 b-values in a single DWI acquisition. For studies on these scanners the four b-value protocol DWI consisted of three consecutive two b-value series ([0,800], [0,600] and [0,100] s/mm2). A separate “merged series multi-b” (MSMB) series containing all b>0 images from the three series plus a single b=0 s/mm2 image created by averaging the three acquired b=0 images was created for analysis and is also included in the collection. The MSMB series are identified by Series Number (0020,0011) ending in 28 and by the string “MSMB” as part of the Series Description (0008,103e). The DICOM metadata fields in the MSMB series have been set to permit reading as a standard GEMS multi-b acquisition. During automated QC procedures, the MSMB acquisitions were rejected if any sequence related parameters other than the b value differed between the multiple two b-value acquisitions. The original 2-b acquisition series are also included in the collection.
For test/retest MRI studies the patient was initially positioned in the scanner and had scout, T2weighted and DWI acquisitions performed. The patient was then removed from the scanner bed and repositioned. The scout, T2weighted and DWI were repeated and then the DCE scans were conducted. All of these acquisitions were stored in a single MRI study for this collection.
Derived object series:
Derived objects from the DWI and DCE acquisitions are included for the convenience of the user. Some of these are not strictly DICOM compliant and may not be readable by all DICOM software packages. Parametric maps and segmentations are believed to be identical to those utilized in the primary analysis and test/retest repeatability analyses of the 6698 Trial, but in some cases modifications may have occurred in subsequent processing.
DWI “Package” for each study with analyzable diffusion scan
- 4 b-value DW Trace Images
- Multiple 2-b MSMB DWI acquisitions were combined (1 site)
- 3 directional DW images were geometrically averaged to generate trace images for studies where directional DWI data was saved
- DICOM enhanced MRI b-value field ( 0018,9087 ) has been populated in all images with the appropriate b-value
- Image order was standardized
- 4-b value ADC maps
- A low threshold (b=0 image > 10) was applied to all studies to suppress background pixels
- ADC was calculated as linear fit to log(S(b)) = log(S(0)) – ADC * b
- Voxels below threshold or with ADC < 0.0 were set to 0
- Manually defined multi-slice tumor segmentations
- ROIs defined for published primary analysis
- Tumor segmented on all slices with identifiable tumor referencing high-b image, ADC map, and DCE subtraction image (for localization)
- The segmentations are provided both as DICOM SEG objects and as DICOM MRI objects on TCIA
These DWI derived series can be identified either by Series Description (0008,103e) or by Series Number (0020,0011). See Table 1 for series identification details.
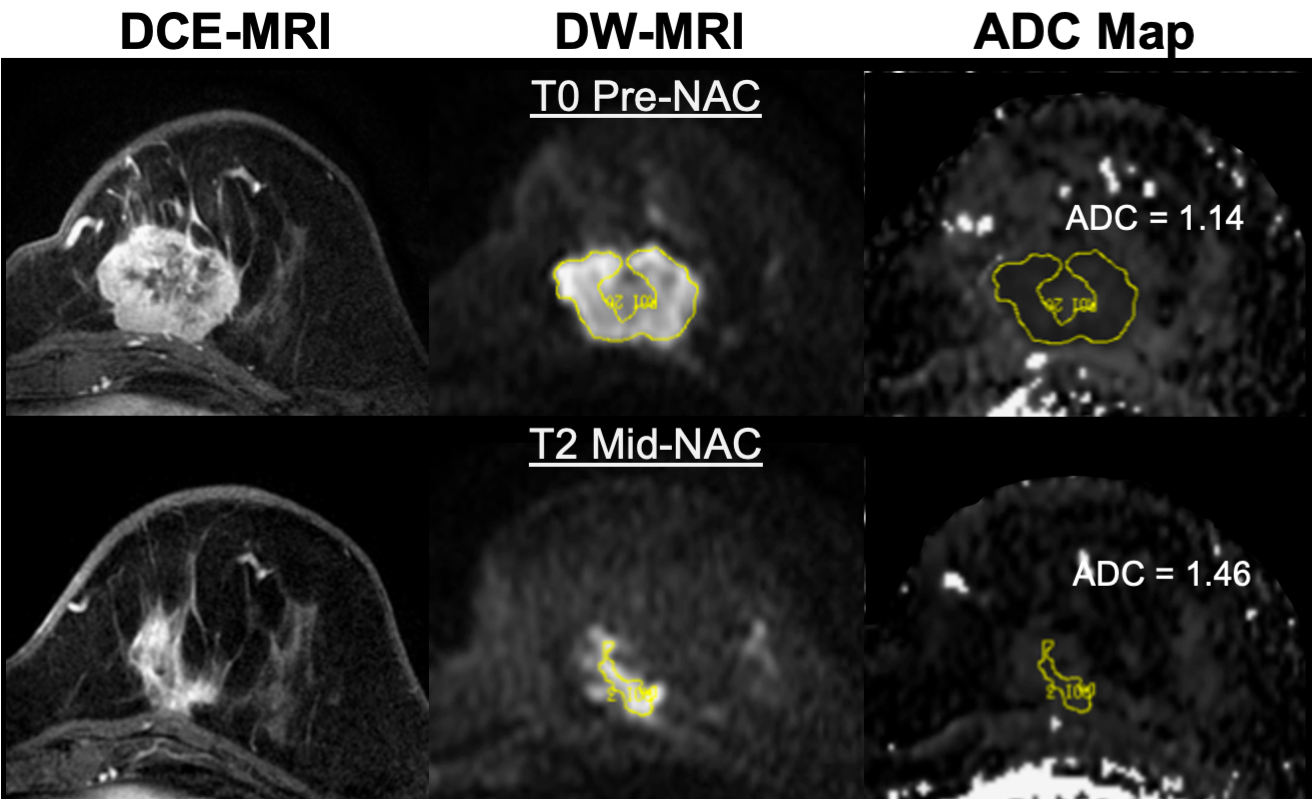
Manual DWI Whole-Tumor Segmentation Method
The segmentations provided in the derived diffusion objects are those generated for and used in the primary analysis and test/retest analyses for the ACRIN 6698 study. Tumor was identified on post-contrast DCE subtraction images and then localized on the ADC map. Multi-slice, whole-tumor regions of interest (ROIs) were manually defined by selecting regions with low ADC and hyperintensity on a high b-value DWI (b=600 or 800 s/mm2) while avoiding adjacent adipose and fibroglandular tissue, biopsy clip artifacts, and regions of high T2 signal (e.g., seroma and necrosis). For multicentric/multifocal disease, all disease regions were included in the ROI. For studies at later treatment timepoints where no tumor could be identified, an ROI was defined in normal appearing fibroglandular tissue in the vicinity of the primary tumor region identified on the patient’s baseline (T0) visit. Region definition was done at the UCSF processing lab using in-house software tools developed with IDL (Exelis Visual Information Solutions, Boulder, Colorado).
DCE “Package” for each study
- Single-breast ipsilateral cropped DCE images
- As this was done primarily for computational efficiency, low matrix-sized images (<384 voxels per row) were stored and processed without cropping
- All maps and segmentations are based on these cropped images
- Enhancement maps
- PEearly: Percent enhancement at effective post-contrast time 120-150 sec
- PElate : Percent enhancement at effective post-contrast time ~450 sec
- SER: Signal enhancement ratio SER = PEearly / PElate
Note: SER values were scaled by 1000 to allow integer storage. DICOM software utilizing the rescale slope and intercept fields [ DICOM fields (0028, 1052-1054)] should return the original SER values in the range from 0.0 – 3.0. - All maps are stored as DICOM MRI objects
- Functional Tumor Volume (FTV) analysis mask
- Image storing FTV masking processes on a pixel-by-pixel basis. The mask encodes:
- Pre-contrast background thresholding
- Minimum PE thresholding (70% nom.)
- Manually defined rectangular volume of interest (VOI) for enhancing tumor analysis
- Manually defined “OMIT” regions used to exclude non-tumor enhancing regions that encroached on the rectangular VOI
- Stored as DICOM SEG object in a separate series (see Table 1)
- Download Analysis mask files description for further information:
- Image storing FTV masking processes on a pixel-by-pixel basis. The mask encodes:
These DCE derived series can be identified either by Series Description (0008,103e) or by Series Number (0020,0011). See Table 1 for series identification details.
Table 1. Series identification for derived DWI and DCE objects
|
Data type |
Series number |
Series Description Match |
| DWI root directory # | Siemens, GE (4b): <root> = Original DWI series number
GE (MSMB): <root> = <1st original series>28 Philips: <root> = Original DWI series number / 100 |
|
|---|---|---|
| TRACE Images | <root>0100 | ACRIN-6698: *DWI TRACE* |
| ADC Maps | <root>0200 | ACRIN-6698: *ADC* |
| Whole tumor mask | <root>9000 | ACRIN-6698: *DWI MASK* |
| Whole tumor segmentation | <root>9001 | ACRIN-6698: *DWI SEG* |
| Merged 2b series (MSMB) | <root> | ACRIN-6698: MergedMSMB* |
| DCE root directory # | Siemens, GE: <root> = 1st original DCE series number
Philips: <root> = Original DCE series number / 100 |
|
| SER Map | <root>1000 | ISPY2: VOLSER: uni-lateral cropped: SER |
| PE phase <n> Map | <root>100<n> | ISPY2: VOLSER: uni-lateral cropped: PE<n> |
| Ipsilateral cropped Original | <root>1800 | ISPY2: VOLSER: uni-lateral cropped: original DCE |
| FTV Analysis Mask | <root>1900 | ISPY2: VOLSER: uni-lateral cropped: Analysis Mask |
- For studies with test/retest DWI scans the prefixes on the series descriptions for the derived DWI objects will be “ACRIN 6698: TrT0:” and “ACRIN 6698: TrT1” for the 1st and 2nd scans respectively.
- FTV Analysis Masks are bit-encoded segmentations encoding all masking steps in the I‑SPY 2 primary FTV analysis. See document Analysis mask files description for a full description of these objects.
- DCE images with less than 384 voxels/row were not cropped for the FTV analysis. For these studies the derived DCE objects are all full bilateral images.
DICOM Dictionary for UCSF Private Attributes in Derived Objects:
Calculation parameters and DCE volume analysis results are included in private attributes in some of the derived objects. ACRIN 6698 ISPY2 Shared Private Tag Data Dictionary and ACRIN 6698 ISPY2 DWI and DCE MRI Data Descriptions files are provided for download for researchers wishing to use these parameters and/or results from the primary analyses. Note: the DCE functional tumor volume (FTV) results stored in the SER derived objects are NOT identical to those used in the I‑SPY 2 study due to differences in implementations on different platforms (UCSF in-house software vs. Hologic Inc. AEGIS system). While the differences are generally small and the AEGIS system was validated against the UCSF results in the prior I‑SPY 1 TRIAL, identical results cannot be guaranteed.
Image Quality Control System
ACRIN 6698 analysis included use of an image quality control system developed specifically for determination of the analyzability of breast DWI for quantitative cancer treatment monitoring. The image quality control system was comprised of three sequential but independent assessment stages: protocol compliance, image quality and usability, and ROI confidence. Since protocol compliance, image quality, and ROI confidence varied between exams for the same patient, each step was performed independently for each MRI exam.
Protocol Compliance Assessment
First, image metadata in DICOM tags was assessed for compliance with specified imaging parameters. Deviations from the specified imaging protocol were documented and classified into minor (non-critical to study aims) and major (critical to the study aims). Images exhibiting major protocol deviations were flagged and omitted from further analysis.
Image Quality and Usability Assessment
Next, the overall image quality and the usability of images for the study endpoints were evaluated. Readers scored image quality using a standard letter grading scale of A (best), B (good), C (average), D (poor) and F (worst) on fat suppression, severity of artifacts (displacement, ghosts, and distortions) and signal-to-noise ratio (SNR). These metrics were selected to reflect the most common and debilitating limitations in breast DWI. Fat suppression and SNR were scored using only the b=0 images while the severity of artifacts was evaluated using all DWI images. Scores were assigned numeric values (A=4 to F=0) and the geometric mean of the scores for fat suppression, artifacts and SNR was computed to yield a composite quality score. Each composite quality score was classified into overall high-, moderate-, or low-quality categories.
Separate from this image quality assessment, a subjective pass/fail assessment was made of image usability for evaluating diffusion parameters such as lesion ADC. This criterion was established to ensure that images of sufficient quality to address the scientific aim of the study were not rejected, e.g., a poor quality score due to excessive imaging artefacts might not result in rejection if the artefacts did not encroach on the region of the enhancing tumor.
ROI Confidence Assessment
After the full multi-slice/multi-region ROIs were placed, they were scored qualitatively based on reader confidence that the ROI faithfully represented the tissue of interest. The goal was to characterize the whole tumor while excluding fat, tissue, and tumor necrosis. ROIs were scored on a numerical scale of 3 for high confidence, 2 for moderate confidence and 1 for low confidence. Inclusion in the primary analysis was determined based on the image quality and ROI confidence assessments.
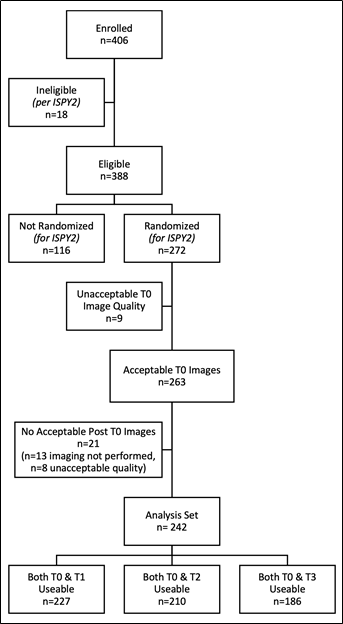
Patient cohorts in the shared collection
Figure 2 shows the enrollment numbers and exclusions used to determine the analysis cohort for the primary analysis. For this TCIA collection we include MRI studies from 385 of the 388 eligible subjects, three being excluded due to having no analyzable MRI studies received by the central imaging lab. The 116 patients excluded from the primary analysis as “Not Randomized” were those screened out of I‑SPY 2, most commonly due to being in the low-risk category (HR+/HER2- subtype with low MammaPrint score). Note that all these patients have baseline MRI studies included in this collection, and some were included in the ACRIN 6698 test/retest repeatability arm. A separate TCIA download option is provided for researchers who wish to focus on the 242-patient cohort used in the primary analysis. These subjects all had MRI studies with DWI scans that passed the QC evaluations at baseline and at at least one later time point. One other download option is provided for the 71-patient cohort with analyzable test/retest, primarily for use in the BMMR2 challenge and for researchers wishing to check reproducibility while focusing on the smaller 242 patient analysis cohort. The primary aim and test/retest studies are the only ones guaranteed to have tumor segmentations and DWI derived objects in this collection.
Figure 2. Derivation of the primary analysis cohort for ACRIN 6698. To be included in the primary analysis patients were required to have analyzable DW-MRI acquisitions at baseline (T0, pre-treatment) and for at least 1 subsequent study time point.
A subset of the ACRIN 6698 collection was used for the NCI QIN sponsored BMMR2 challenge (Breast Multiparametric MRI for prediction of NAC Response) running from June 1, 2021 through December 21, 2021. The goal of the challenge was to identify image-based markers derived from DW-MRI, alone or in combination with DCE-MRI, with improved performance over whole-tumor mean ADC for predicting pCR following NAC for invasive breast cancer. To achieve this goal, a subset of the ACRIN 6698 MRI studies from 191 subjects with analyzable data at all of the 1st three timepoints (T0, T1 and T2) was made available to registered research teams for the sole purpose of participating in the challenge. The cohort was split 60%/40% into training and test groups with pCR outcome data provided for the training set only. Download manifests for the challenge groups are provided here to allow direct comparison of future analyses with the challenge results.
Processing and transfer history for MRI studies in this collection
The processing of the MR image data for this ACRIN 6698 collection consisted of the following steps between image acquisition and the creation of this shared data set on TCIA:
- MRI studies were conducted on approved scanners and were transferred to a system on-site for further processing. In some cases, this may have included an intermediate transfer to the site’s PACs system.
- MRI studies were sent from the study centers to the ACRIN Core Lab using the ACR TRIAD program
- Image data were de-identified and centrally archived at the ACRIN Core Lab
- Archived data was downloaded to the Breast Imaging Research Program (BIRP) at the University of California, San Francisco (UCSF) for DWI ADC analysis and DCE volumetric analysis
- De-identified image data, derived analysis maps and segmentations, and ancillary data files were transferred from UCSF to TCIA for data sharing
While every effort was made to preserve the integrity of both the original image data and image meta-data (DICOM attributes, public and private), multiple file transfers and strict adherence to HIPAA guidelines for patient confidentiality may have resulted in loss of some data.
Ancillary files for the ACRIN 6698 collection
The following links will download files with non-image information for this collection.
BMMR2 Challenge patient demographic, clinical and outcome data files for the training and test cohorts (comma-separated text files).
Due to the ongoing nature of the I‑SPY 2 Trial only this limited set of patient data can be released at this time. Further information will become available with release of further I‑SPY 2 imaging data.
BMMR2 Training set cases with clinical data
BMMR2 Testing set cases with clinical data
ACRIN 6698 test-retest cases with clinical data
Full collection ancillary patient information file (Excel workbook file). Includes clinical and outcome data, DWI Quality Control ratings, and cohort identification.
Full Collection Ancillary Patient Information file
ACRIN 6698 MRI protocol parameter specifications
ACRIN 6698 MRI Acquisition Parameters
DCE analysis mask description. This file provides usage information for the functional tumor volume analysis masks provided as part of the DCE derived objects package.
Analysis mask files description
DICOM Dictionary and data descriptions for UCSF derived objects (Excel and pdf files)
ACRIN 6698 ISPY2 Shared Private Tag Data Dictionary
ACRIN 6698 ISPY2 DWI and DCE MRI Data Descriptions
Acknowledgements
We would like to acknowledge the individuals and institutions that have made this collection possible.
- The patients who have volunteered to participate in the ACRIN 6698 and I-SPY 2 Trials
- The extensive network of I-SPY 2 investigators, patient advocates and study coordinators.
- I-SPY 2 site radiology teams that have contributed substantially to the value of this archive.
- Support from the NIH including grants to UCSF (U01 CA225427, R01 CA132870) and ACRIN (U01 CA079778, U01 CA080098).
- The I-SPY 2 TRIAL is supported by the Quantum Leap Healthcare Collaborative.
Related Publications
Publications by the Dataset Authors
The authors recommended the following as the best source of additional information about this dataset:
Publication Citation |
|
|
Newitt, D. C., Zhang, Z., Gibbs, J. E., Partridge, S. C., Chenevert, T. L., Rosen, M. A., Bolan, P. J., Marques, H. S., Aliu, S., Li, W., Cimino, L., Joe, B. N., Umphrey, H., Ojeda‐Fournier, H., Dogan, B., Oh, K., Abe, H., Drukteinis, J., … Esserman, L. J. (2018). Test–retest repeatability and reproducibility of ADC measures by breast DWI: Results from the ACRIN 6698 trial. Journal of Magnetic Resonance Imaging, 49(6), 1617–1628. https://doi.org/10.1002/jmri.26539 |
Publication Citation |
|
|
Partridge, S. C., Zhang, Z., Newitt, D. C., Gibbs, J. E., Chenevert, T. L., Rosen, M. A., Bolan, P. J., Marques, H. S., Romanoff, J., Cimino, L., Joe, B. N., Umphrey, H. R., Ojeda-Fournier, H., Dogan, B., Oh, K., Abe, H., Drukteinis, J. S., Esserman, L. J., & Hylton, N. M. (2018). Diffusion-weighted MRI Findings Predict Pathologic Response in Neoadjuvant Treatment of Breast Cancer: The ACRIN 6698 Multicenter Trial. Radiology, 289(3), 618–627. https://doi.org/10.1148/radiol.2018180273 |
No other publications were recommended by dataset authors.
Research Community Publications
TCIA maintains a list of publications that leveraged this dataset. If you have a manuscript you’d like to add please contact TCIA’s Helpdesk.