HNSCC-mIF-mIHC-comparison | AI-ready restained and co-registered multiplex dataset for head-and-neck carcinoma
DOI: 10.7937/TCIA.2020.T90F-WB82 | Data Citation Required | 2k Views | Image Collection
| Location | Species | Subjects | Data Types | Cancer Types | Size | Status | Updated | |
|---|---|---|---|---|---|---|---|---|
| Head-Neck | Human | 8 | Histopathology | Head and Neck Cancer | Image Analyses, Software/Source Code | Public, Complete | 2023/08/31 |
Summary
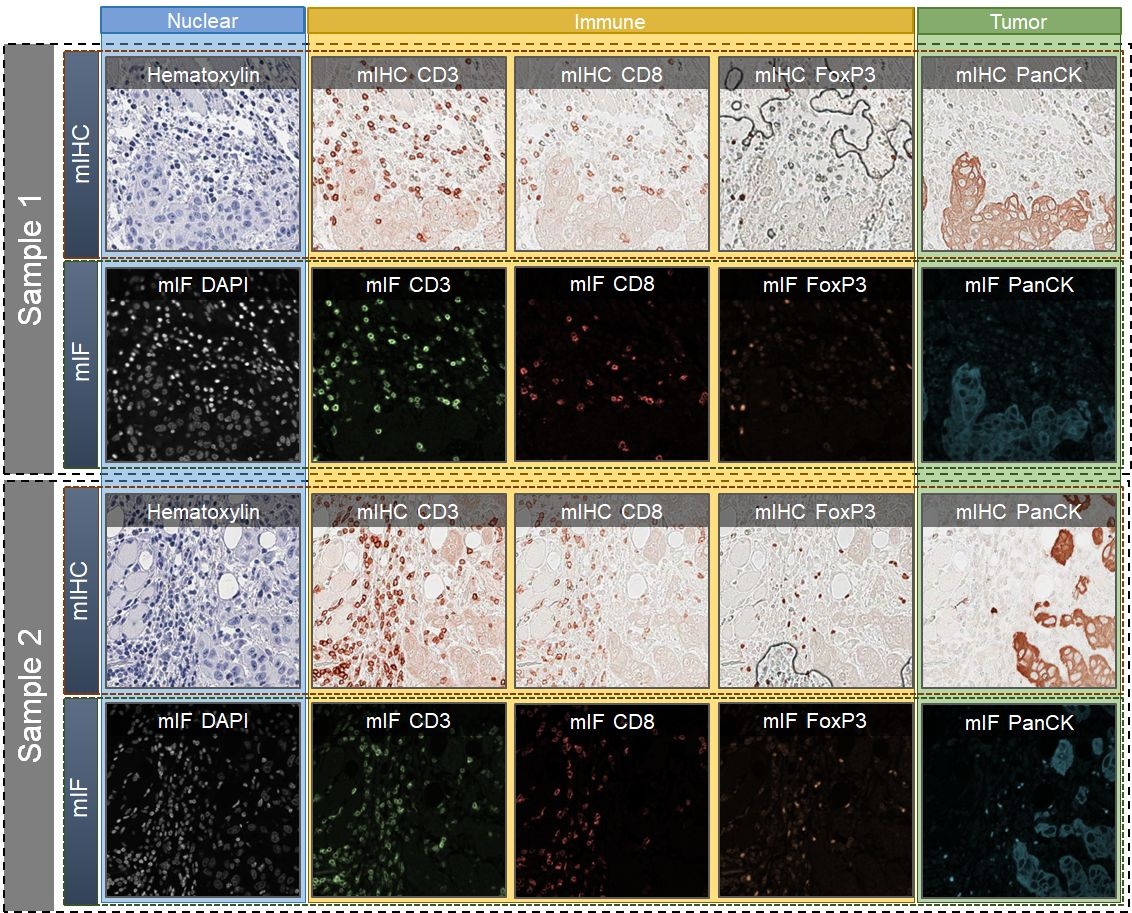
We introduce a new AI-ready computational pathology dataset containing restained and co-registered digitized images from eight head-and-neck squamous cell carcinoma patients. Specifically, the same tumor sections were stained with the expensive multiplex immunofluorescence (mIF) assay first and then restained with cheaper multiplex immunohistochemistry (mIHC). This is a first public dataset that demonstrates the equivalence of these two staining methods which in turn allows several use cases; due to the equivalence, our cheaper mIHC staining protocol can offset the need for expensive mIF staining/scanning which requires highly skilled lab technicians. As opposed to subjective and error-prone immune cell annotations from individual pathologists (disagreement > 50%) to drive SOTA deep learning approaches, this dataset provides objective immune and tumor cell annotations via mIF/mIHC restaining for more reproducible and accurate characterization of tumor immune microenvironment (e.g. for immunotherapy). We demonstrate the effectiveness of this dataset in three use cases: (1) IHC quantification of CD3/CD8 tumor-infiltrating lymphocytes via style transfer, (2) virtual translation of cheap mIHC stains to more expensive mIF stains, and (3) virtual tumor/immune cellular phenotyping on standard hematoxylin images. The code for stain translation is available at https://github.com/nadeemlab/DeepLIIF and the code for performing interactive deep learning whole-cell/nuclear segmentation is available at https://github.com/nadeemlab/impartial. After scanning the full images, nine regions of interest (ROIs) from each slide/Case were chosen by an experienced pathologist on both mIF and mIHC images: three in the tumor core (T), three at the tumor margin (M),and three outside in the adjacent stroma (S) area. These individual ROIs were further subdivided into four 512x512 patches with indices [0_0], [0_1], [1_0], [1_1]. The final notation for each file is Case[patient_id]_[T/M/S][1/2/3]_[ROI_index]_[Marker_name]. More details can be found in the paper.
Data Access
Version 2: Updated 2023/08/31
Version 2 dataset modifications:
(1) 35 channels by human error in conversion in the version 1 dataset were corrected.
(2) Non-standard im3 format, that is not supported by most platforms/viewers, images were replaced with png format.
(3) A lot of images in the multiplex IHC folder were not from the same ROI as the hematoxylin/AEC. Names/labels for all the files were corrected to address this.
(4) Grayscale images which do not allow to analyze the original AEC/Hematoxylin colored images, so original-colored images were added.
(5) Intensity concordance study was difficult with the old version since the images across AEC/mpIF were not perfectly co-registered. Images are now perfectly co-registered to address this.
(6) The original focus was not on the AI-ready datasets. In this version, we release an AI-ready dataset that should work out-of-the-box for multiple tasks using the SOTA deep learning algorithms.
| Title | Data Type | Format | Access Points | Subjects | License | Metadata | |||
|---|---|---|---|---|---|---|---|---|---|
| Tissue Slide Images | Histopathology | PNG | Download requires IBM-Aspera-Connect plugin |
8 | 3,216 | CC BY 4.0 | — |
Additional Resources for this Dataset
- The source code for stain translation is available at https://github.com/nadeemlab/DeepLIIF
- The source code for performing interactive deep learning whole-cell/nuclear segmentation is available at https://github.com/nadeemlab/impartial
Citations & Data Usage Policy
Data Citation Required: Users must abide by the TCIA Data Usage Policy and Restrictions. Attribution must include the following citation, including the Digital Object Identifier:
Data Citation |
|
|
Ghahremani, P., Marino, J., Hernandez-Prera, J., de la Iglesia, J. V., Slebos, R. J., Chung, C. H., & Nadeem, S. (2023). AI-ready re-stained and co-registered multiplex dataset for head-and-neck carcinoma (HNSCC-mIF-mIHC-comparison) (Version 2) [dataset]. The Cancer Imaging Archive. https://doi.org/10.7937/TCIA.2020.T90F-WB82 |
Detailed Description
Version 2 of dataset replaced title, summary, acknowledgements, and publication citation with new information. These entries for version 1 dataset may be accessed here.
Acknowledgements
This work was supported by MSK Cancer Center Support Grant/Core Grant (P30 CA008748) and by James and Esther King Biomedical Research Grant (7JK02) and Moffitt Merit Society Award to C. H. Chung. It is also supported in part by the Moffitt’s Total Cancer Care Initiative, Collaborative Data Services, Biostatistics and Bioinformatics, and Tissue Core Facilities at the H. Lee Moffitt Cancer Center and Research Institute, an NCI-designated Comprehensive Cancer Center (P30-CA076292).
Related Publications
Publications by the Dataset Authors
The authors recommended the following as the best source of additional information about this dataset:
Publication Citation |
|
|
Ghahremani, P., Marino, J., Hernandez-Prera, J., de la Iglesia, J. V., Slebos, R. J., Chung, C. H., & Nadeem, S. (2023). An AI-Ready Multiplex Staining Dataset for Reproducible and Accurate Characterization of Tumor Immune Microenvironment. In: H. Greenspan et al. (eds.): Medical Image Computing and Computer Assisted Intervention – MICCAI 2023. MICCAI 2023. Lecture Notes in Computer Science, vol 14225, pp. 1–10, 2023. Springer, Cham. https://doi.org/10.1007/978-3-031-43987-2_68. |
No other publications were recommended by dataset authors.
Research Community Publications
TCIA maintains a list of publications that leveraged this dataset. If you have a manuscript you’d like to add please contact TCIA’s Helpdesk.
Previous Versions
Version 1: Updated 2020/06/04
Version 2 of dataset replaced title, summary, acknowledgements, and publication citation with new information. These entries for version 1 dataset may be accessed here.
| Title | Data Type | Format | Access Points | License | Metadata | ||||
|---|---|---|---|---|---|---|---|---|---|
| Tissue Slide Images | IM3 and TIFF | Download requires IBM-Aspera-Connect plugin |
— |
