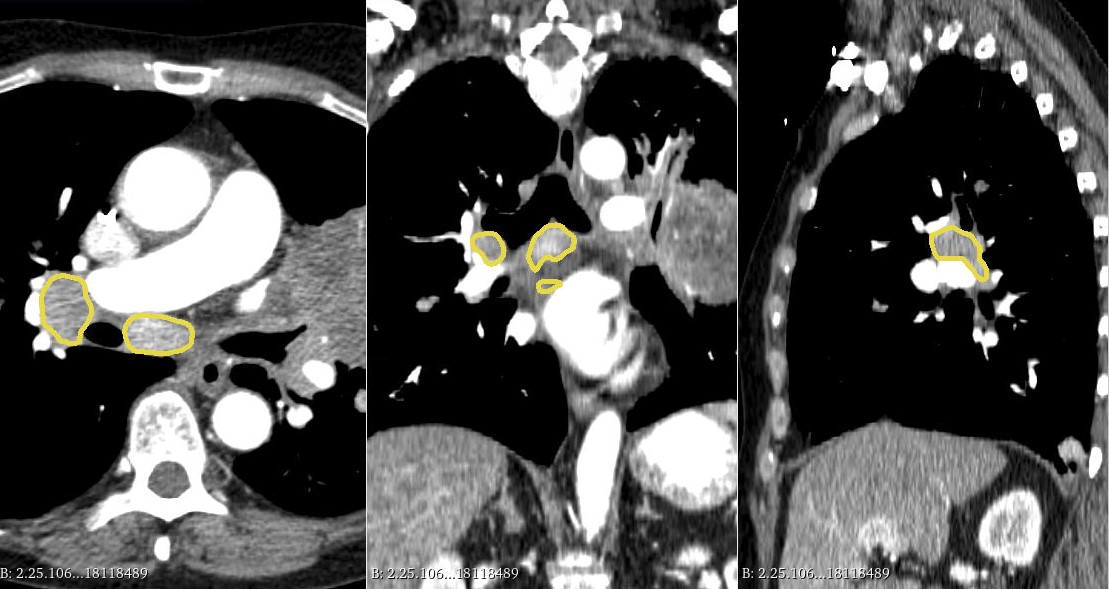
Mediastinal-Lymph-Node-SEG | Mediastinal Lymph Node Quantification (LNQ): Segmentation of Heterogeneous CT Data
DOI: 10.7937/qvaz-ja09 | Data Citation Required | 6k Views | Image Collection
| Location | Species | Subjects | Data Types | Cancer Types | Size | Status | Updated | |
|---|---|---|---|---|---|---|---|---|
| Lymph Node | Human | 513 | Demographic, Diagnosis, CT, SEG | Various, Breast Cancer, Non-small Cell Lung Cancer, Hodgkin Lymphoma, Small Cell Lung Cancer, Thyroid Cancer, Adenocarcinoma, Melanoma, Head and Neck Cancer, Prostate Cancer, Mesothelioma, Ovarian Cancer, Colon Cancer | Clinical | Public, Complete | 2024/08/28 |
Summary
The dataset concerns patients with lymphadenopathy (i.e., swelling of lymph nodes) due to illness or disease, such as cancer or infections. Lymph node disease involvement is due to the body’s immune response. The cohort is a cross-institutional dataset of chest CT scans acquired from 513 patients during treatment for various cancer types, curated for the LNQ2023 MICCAI Challenge to help develop new segmentation tools from weakly annotated cases. The dataset originates from the Tumor Imaging Metrics Core (TIMC), a multi-institutional imaging core lab (Massachusetts General Hospital, Dana Farber Cancer Institute, Brigham and Women’s Hospital, Boston, MA, USA) that provides multimodality imaging measurements to evaluate treatment response in patients enrolled in oncology clinical trials. TIMC has processed data from over fourteen hundred clinical trials conducted in the period 2007- 2020. The dataset comprises patients with various types of cancer. The top six primary patient cancers which accounted for 60% of all patients were: breast cancer, chronic lymphocytic leukemia (CLL), lung non-small cell cancer, Hodgkin‘s lymphoma, small cell lung cancer, and renal cell cancer. The remaining patient cancer types accounted for 40% of all patients including thyroid cancer, adenocarcinoma, endometrial adenocarcinoma, melanoma, head and neck cancer, non-Hodgkin’s lymphoma, prostate cancer, mesothelioma, esophageal cancer, ovarian cancer, and colon cancer. All of the scans in the dataset were captured at baseline timepoints during clinical trials. The patients have likely been through standard of care before being placed on the trials, and may have even been through more than one trial. The patient's sex and primary cancer type are also provided. These patients were being treated in clinical trials for their conditions and have therefore undergone pathology review and other eligibility checks prior to enrollment. All annotators were trained radiologists or radiology domain experts with over ten years of experience. The initial localization for the lymph nodes in our partially annotated cases was performed as part of the clinical trial reads by the staff at the Tumor Imaging Metrics Core (TIMC), and each case was read by US-board certified radiologists in addition to TIMC Image Analysts. Annotators on our project extended the initial localizations into full segmentations in a subset of cases. Volumetric measurements were performed using 3D Slicer (Pieper et al., 2004; Fedorov et al., 2012). For each patient, the CT scan as well as the bidimensional measurements of the target lymph node were loaded into 3D Slicer (version 4.11), and then the Segment Editor module was used to perform manual delineation of the lymph node boundary. The Draw tool within the Editor module was used to draw free hand boundaries on axial cross-sections while viewing the sagittal and coronal planes for reference. Excluded from the lesion boundary were large vessels, artifacts, and non-nodal components.
Data Access
Version 1: Updated 2024/08/28
| Title | Data Type | Format | Access Points | Subjects | License | Metadata | |||
|---|---|---|---|---|---|---|---|---|---|
| Images | CT, SEG | DICOM | Download requires NBIA Data Retriever |
513 | 513 | 1,026 | 66,870 | CC BY 4.0 | View |
| Clinical Data | Demographic, Diagnosis | CSV | 513 | CC BY 4.0 | — |
Additional Resources for this Dataset
The NCI Cancer Research Data Commons (CRDC) provides access to additional data and a cloud-based data science infrastructure that connects data sets with analytics tools to allow users to share, integrate, analyze, and visualize cancer research data.
- Imaging Data Commons (IDC) (Imaging Data)
Citations & Data Usage Policy
Data Citation Required: Users must abide by the TCIA Data Usage Policy and Restrictions. Attribution must include the following citation, including the Digital Object Identifier:
Data Citation |
|
|
Idris, T., Somarouthu, S., Jacene, H., LaCasce, A., Ziegler, E., Pieper, S., Khajavi, R., Dorent, R., Pujol, S., Kikinis, R., & Harris, G. (2024). Mediastinal Lymph Node Quantification (LNQ): Segmentation of Heterogeneous CT Data (Version 1) [Data set]. The Cancer Imaging Archive. https://doi.org/10.7937/QVAZ-JA09 |
Acknowledgements
The Brigham and Women’s Hospital provided funding for this dataset through NIH grant “Lymph Node Quantification System for Multisite Clinical Trials” (5R01CA235589).
Related Publications
Publications by the Dataset Authors
The authors recommended the following as the best source of additional information about this dataset:
No other publications were recommended by dataset authors.
Research Community Publications
TCIA maintains a list of publications that leveraged this dataset. If you have a manuscript you’d like to add please contact TCIA’s Helpdesk.