QIN-HEADNECK | QIN-HEADNECK
DOI: 10.7937/K9/TCIA.2015.K0F5CGLI | Data Citation Required | 2.4k Views | 23 Citations | Image Collection
| Location | Species | Subjects | Data Types | Cancer Types | Size | Status | Updated | |
|---|---|---|---|---|---|---|---|---|
| Head-Neck | Human | 279 | Demographic, Exposure, Treatment, Diagnosis, Follow-Up, CT, PT, SR, SEG, RWV | Head and Neck Carcinomas | Clinical, Image Analyses | Limited, Complete | 2020/09/15 |
Summary
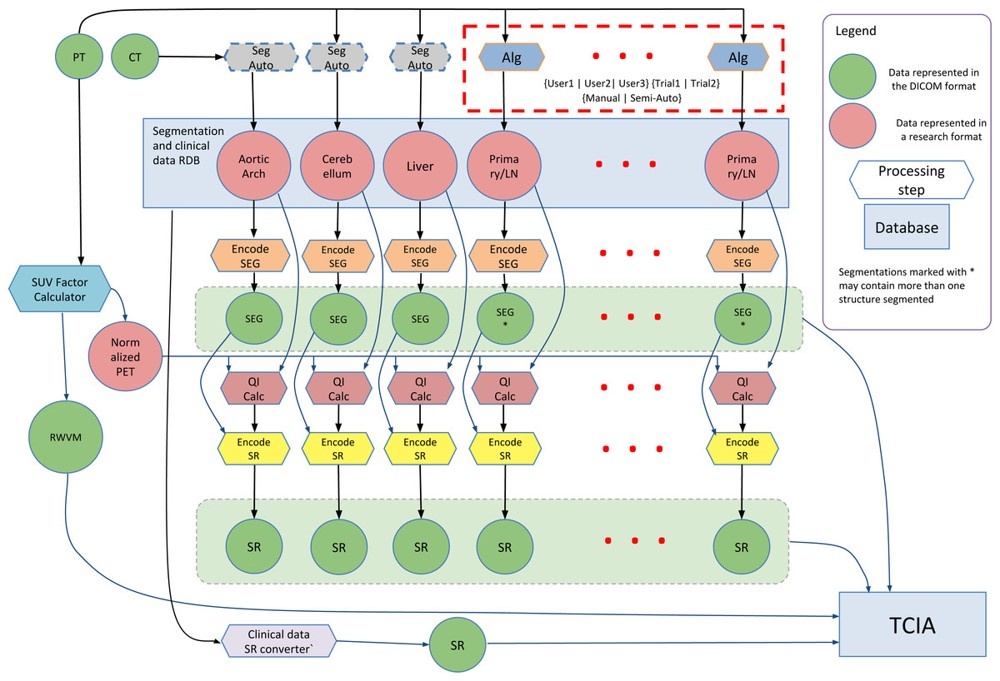
This collection is a set of head and neck cancer patients' multiple positron emission tomography/computed tomography (PET/CT) 18F-FDG scans–before and after therapy–with follow up scans where clinically indicated. The data was provided to help facilitate research activities of the National Cancer Institute's (NCI's) Quantitative Imaging Network (QIN). This collection was supported by Grants: U24 CA180918 (http://qiicr.org) and U01 CA140206. The following schematic summarizes much of the work done within the QIICR grant to augment the PET/CT scans with segmentations and clinical data using the DICOM standard: (click to enlarge) The mission of the QIN is to improve the role of quantitative imaging for clinical decision making in oncology by developing and validating data acquisition, analysis methods, and tools to tailor treatment for individual patients and predict or monitor the response to drug or radiation therapy. More information is available on the Quantitative Imaging Network Collections page. Interested investigators can apply to the QIN at: Quantitative Imaging for Evaluation of Responses to Cancer Therapies (U01) PAR-11-150.About the NCI QIN
Data Access
Some data in this collection contains images that could potentially be used to reconstruct a human face. To safeguard the privacy of participants, users must sign and submit a TCIA Restricted License Agreement to help@cancerimagingarchive.net before accessing the data.
Version 4: Updated 2020/09/15
Added 123 new subjects (Patient IDs = QIN-HeadNeck-02-####). Added missing PT or CT pre-treatment and follow up scans to 28 of the previously existing QIN-HeadNeck-01-#### subjects. Added supporting clinical data in XLSX format for all patients.
| Title | Data Type | Format | Access Points | Subjects | License | Metadata | |||
|---|---|---|---|---|---|---|---|---|---|
| Images and Segmentations | CT, PT, SR, SEG, RWV | DICOM | 279 | 1,032 | 3,837 | 701,002 | NIH Controlled Data Access Policy | View | |
| Clinical Data (See also Detailed Description) | Demographic, Exposure, Treatment, Diagnosis, Follow-Up | XLSX | CC BY 3.0 | — |
Citations & Data Usage Policy
Data Citation Required: Users must abide by the TCIA Data Usage Policy and Restrictions. Attribution must include the following citation, including the Digital Object Identifier:
Data Citation |
|
|
Beichel, R. R., Ulrich, E. J., Bauer, C., Wahle, A., Brown, B., Chang, T., Plichta, K., Smith, B., Sunderland, J., Braun, T., Fedorov, A., Clunie, D., Onken, M., Magnotta, V. A., Menda, Y., Riesmeier, J., Pieper, S., Kikinis, R., Graham, M.M., Casavant T. L., Sonka M,. & Buatti, J. (2015). Data From QIN-HEADNECK (Version 4) [Data set]. The Cancer Imaging Archive. https://doi.org/10.7937/K9/TCIA.2015.K0F5CGLI |
Detailed Description
Associated Clinical Metadata
- Structured Report DICOM objects (Modality SR), are available for a subset of these subjects in the DICOM downloads and can be distinguished from image files by the series description “Clinical Data.” Note, there is no image preview thumbnail for a Structured Report.
Related Publications
Publications by the Dataset Authors
The authors recommended the following as the best source of additional information about this dataset:
Publication Citation |
|
|
Fedorov, A., Clunie, D., Ulrich, E., Bauer, C., Wahle, A., Brown, B., Onken, M., Riesmeier, J., Pieper, S., Kikinis, R., Buatti, J., & Beichel, R. R. (2016). DICOM for quantitative imaging biomarker development: a standards based approach to sharing clinical data and structured PET/CT analysis results in head and neck cancer research. In PeerJ (Vol. 4, p. e2057). PeerJ. https://doi.org/10.7717/peerj.2057 |
No other publications were recommended by dataset authors.
Research Community Publications
TCIA maintains a list of publications that leveraged this dataset. If you have a manuscript you’d like to add please contact TCIA’s Helpdesk.
Previous Versions
Version 3: Updated 2019/07/24
Lifted restriction from SR object data download.
| Title | Data Type | Format | Access Points | License | Metadata | ||||
|---|---|---|---|---|---|---|---|---|---|
| Images | CT, PT, SR, SEG, RWV | DICOM | 156 | 651 | 2,993 | 353,408 | — | ||
| DICOM Metadata Digest | CSV | — |
Version 2: Updated 2017/12/06
Downloads require the NBIA Data Retriever .
Added associated DICOM SEG, SR, and RWV objects
| Title | Data Type | Format | Access Points | License | Metadata | ||||
|---|---|---|---|---|---|---|---|---|---|
| Images | CT, PT, SR, SEG, RWV | DICOM | 156 | 495 | 2,837 | 353,252 | — | ||
| DICOM Metadata Digest | CSV | — |