LDCT-and-Projection-data | Low Dose CT Image and Projection Data
DOI: 10.7937/9NPB-2637 | Data Citation Required | 16.9k Views | 62 Citations | Image Collection
| Location | Species | Subjects | Data Types | Cancer Types | Size | Status | Updated | |
|---|---|---|---|---|---|---|---|---|
| Head, Chest, and Abdomen | Human | 299 | CT, Other, Protocol, Demographic, Diagnosis, Measurement | Various | Clinical, Software/Source Code | Limited, Complete | 2023/04/03 |
Summary
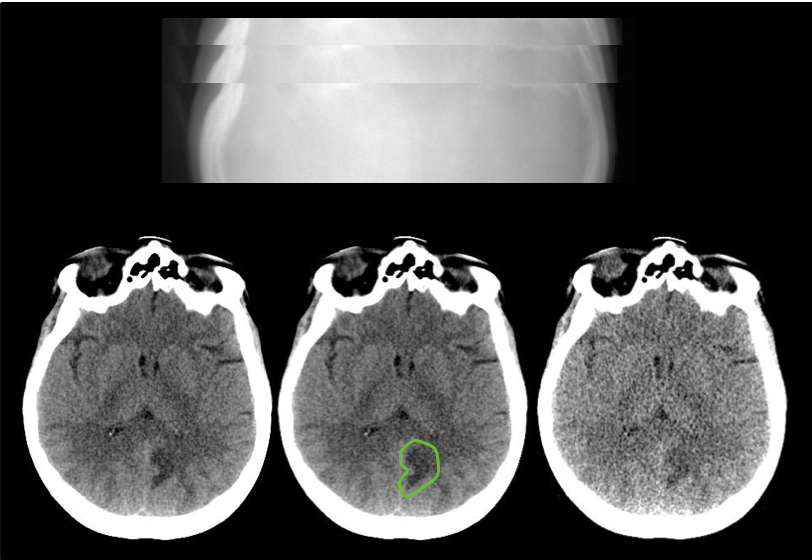
Investigators at the Mayo Clinic, with funding from the National Institute of Biomedical Imaging and Bioengineering (EB 017095 and EB 017185), have built a library of CT patient projection data in an open and vendor-neutral format. This format, referred to as DICOM-CT-PD (Additional information regarding the CT projection data format in the article by Chen et al at doi: 10.1118/1.4935406), is an extended DICOM format that contains CT projection data and acquisition geometry. The de-identified patient projection data in the library were decoded with help of the manufacturer and have been converted into an open standardized format. Reconstructed images, patient age and gender, and pathology annotation are also provided for these de-identified data sets. The library consists of scans from various exam types, including non-contrast head CT scans acquired for acute cognitive or motor deficit, low-dose non-contrast chest scans acquired to screen high-risk patients for pulmonary nodules, and contrast-enhanced CT scans of the abdomen acquired to look for metastatic liver lesions. 2016 Low Dose CT Grand Challenge The 2016 Low Dose CT Grand Challenge, sponsored by the AAPM, NIBIB, and Mayo Clinic, used 30 contrast-enhanced abdominal CT patient scans, 10 for training and 20 for testing. Thirteen of the 20 testing datasets from the Grand Challenge were subsequently included in this larger collection of CT image and projection data (TCIA LDCT-and-Projection-data). Because of the frequency of requests received by Mayo and the AAPM for the complete 2016 Grand Challenge dataset, on September 21, 2021 all 30 cases were updated to use the same projection data format as used for the TCIA data library and made publicly available in a single location. Please refer to the READ ME file at that location for a mapping between the case ID numbers used in the 2016 Grand Challenge and the case ID numbers used in the TCIA library for the 13 cases that exist in both libraries. Additional information about the 2016 Low Dose CT Grand Challenge can be found on the AAPM website and in the Medical Physics paper by McCollough et al.
Data Access
Some data in this collection contains images that could potentially be used to reconstruct a human face. To safeguard the privacy of participants, users must sign and submit a TCIA Restricted License Agreement to help@cancerimagingarchive.net before accessing the data.
Version 6: Updated 2023/04/03
Note the following corrections were made to the clinical data:
- Lesion Z locations for all GE cases now reflect slice number
| Title | Data Type | Format | Access Points | Subjects | License | Metadata | |||
|---|---|---|---|---|---|---|---|---|---|
| Images | CT | DICOM | 299 | 597 | 1,045 | 13,013,532 | NIH Controlled Data Access Policy | View | |
| Images Phantom Object Only | CT | DICOM | 1 | 1 | 1 | 18,032 | NIH Controlled Data Access Policy | View | |
| DICOM-CT-PD User Manual Version 3 | Other, Protocol | CC BY 3.0 | — | ||||||
| Matlab DICOM-CTPD data dictionary | Other | TXT | CC BY 3.0 | — | |||||
| Matlab DICOM-CTPD reader script | Other | MATLAB and ZIP | CC BY 3.0 | — | |||||
| Clinical Data | Demographic, Diagnosis, Measurement | XLSX and ZIP | CC BY 3.0 | — |
Additional Resources for this Dataset
- The Helix2Fan: Helical to fan-beam CT geometry rebinning and differentiable reconstruction of DICOM-CT-PD projections repository on Github provides source code to load raw helical DICOM-CT-PD CT projections and rebin them to flat detector fan-beam geometry.
Citations & Data Usage Policy
Data Citation Required: Users must abide by the TCIA Data Usage Policy and Restrictions. Attribution must include the following citation, including the Digital Object Identifier:
Data Citation |
|
|
McCollough, C., Chen, B., Holmes III, D. R., Duan, X., Yu, Z., Yu, L., Leng, S., & Fletcher, J. (2020). Low Dose CT Image and Projection Data (LDCT-and-Projection-data) (Version 6) [Data set]. The Cancer Imaging Archive. https://doi.org/10.7937/9NPB-2637 |
Acknowledgement |
|
|
Presentations and publications shall acknowledge grants EB017095 and EB017185 (Cynthia McCollough, PI) from the National Institute of Biomedical Imaging and Bioengineering. |
Detailed Description
For each patient CT scan, three types of data are provided: DICOM-CT-PD projection data, DICOM image data, and Excel clinical data reports. CT projection data are provided for both full and simulated lower dose levels and CT image data reconstructed using the commercial CT system are provided for the full dose projection data. For patients scanned on the SOMATOM Definition Flash CT scanner from Siemens Healthcare, CT image data reconstructed using the commercial CT system are also provided for the lower dose projection data. All CT images were reconstructed using a filtered back projection method. Several instructional documents are provided to help users extract needed information from the DICOM-CT-PD files, including a dictionary file for the DICOM-CT-PD format, a DICOM-CT-PD reader, and a user manual.
This collection comprises 99 head scans (labeled N for neuro), 100 chest scans (labeled C for chest), and 100 abdomen scans (labeled L for liver). Fifty cases for each scan type are from a SOMATOM Definition Flash CT scanner (Siemens Healthcare, Forchheim, Germany). Forty-nine head cases, 50 chest cases, and 50 abdomen cases are from a Lightspeed VCT CT scanner (GE Healthcare, Waukesha, WI). Together, these data will greatly facilitate the development and validation of new CT reconstruction and/or denoising algorithms, including those associated with machine learning or artificial intelligence.
Acquisition protocol
All CT scans were acquired at routine dose levels for the practice at which they were obtained using standard-clinical protocols for the anatomical region of interest. Each clinical case was processed to include a second projection dataset at a simulated lower dose level. Head and abdomen cases are provided at 25% of the routine dose and chest cases are provided at 10% of the routine dose.
1Additional information regarding the CT projection data format: Chen B, Duan X, Yu Z, Leng S, Yu L, McCollough CH. Technical Note: Development and validation of an open data format for CT projection data. Med Phys. 2015;42(12):6964. (doi: https://doi.org/10.1118/1.4935406.)
Acknowledgements
This work would not have been possible without the support and efforts of many individuals and organizations.
- A complete list of acknowledgements can be found here.
Related Publications
Publications by the Dataset Authors
The authors recommended the following as the best source of additional information about this dataset:
Publication Citation |
|
|
Moen, T. R., Chen, B., Holmes, D. R., III, Duan, X., Yu, Z., Yu, L., Leng, S., Fletcher, J. G., & McCollough, C. H. (2020). Low dose CT image and projection dataset. Medical Physics. https://doi.org/10.1002/mp.14594 |
No other publications were recommended by dataset authors.
Research Community Publications
TCIA maintains a list of publications that leveraged this dataset. If you have a manuscript you’d like to add please contact TCIA’s Helpdesk.
Previous Versions
Version 5: Updated 2022/12/15
Note: The following corrections were made to the clinical data:
- Incorrect Lesion coordinates
- In some places the x/y coordinates for lesions was incorrect
- In almost all Siemens datasets, the Z axis was flipped
- Incorrect UID
- Correction has been made to case L210 UID
| Title | Data Type | Format | Access Points | License | Metadata | ||||
|---|---|---|---|---|---|---|---|---|---|
| Images | CT | DICOM | Download requires NBIA Data Retriever |
299 | 597 | 1,045 | 13,013,532 | NIH Controlled Data Access Policy | — |
| Images Phantom Object Only | CT | DICOM | Download requires NBIA Data Retriever |
1 | 1 | 1 | 18,032 | NIH Controlled Data Access Policy | — |
| DICOM-CT-PD User Manual Version 3 | CC BY 3.0 | — | |||||||
| Matlab DICOM-CTPD data dictionary | TXT | CC BY 3.0 | — | ||||||
| Matlab DICOM-CTPD reader script | MATLAB and ZIP | CC BY 3.0 | — | ||||||
| Clinical Data | ZIP and CSV | CC BY 3.0 | — |
Version 4: Updated 2022/03/31
Update on the GE data
The following corrections/changes have been made to the GE projection data:
- The value in DICOM tag (0028,1052) RescaleIntercept has been re-calculated and updated.
- DICOM tag (7033,1065) PhotonStatistics, which describes the incident x-ray beam profile after passing through the bowtie filter, has been added for every projection. The beam profile is characterized in terms of noise equivalent number of incident photons (i.e., noise equivalent quanta)..
- DICOM tag (0018,1151) XRayTubeCurrent has been updated for the chest and abdomen cases, which use tube current modulation. The value reflects the tube current for each projection view. With tube current modulation on, this value will vary across different project views. Tube current modulation is off for the head exams.
- It was discovered that the projection data for chest case C197 was a duplication of case C009. Therefore, C197 has been omitted from the dataset, resulting in a total of 49 chest cases.
- The projection data for the sequential head scans are contained in one directory for each dose level for each patient. DICOM tag (0020, 0012) Acquisition Number, which is a number identifying the single continuous gathering of data over a period of time (i.e., one scan), can be used to sort the data based on each sequential acquisition within the exam. A MATLAB code that will sort the data in this manner has been provided here. The .m files must be placed in the same directory containing the projection files to correctly identify and sort them.
The previous version of the GE data has been replaced with the new data (Version 4); click here to download a spreadsheet that maps the old UIDs to the new UIDs. You can access the updated series by downloading this manifest. Please contact the TCIA Helpdesk at help@cancerimagingarchive.net with any questions.
| Title | Data Type | Format | Access Points | License | Metadata | ||||
|---|---|---|---|---|---|---|---|---|---|
| Images | CT | DICOM | Download requires NBIA Data Retriever |
299 | 597 | 1,045 | 13,013,532 | — | |
| Images Phantom Object Only | CT | DICOM | Download requires NBIA Data Retriever |
1 | 1 | 1 | 18,032 | — | |
| DICOM-CT-PD User Manual Version 3 | — | ||||||||
| Matlab DICOM-CTPD data dictionary | TXT | — | |||||||
| Matlab DICOM-CTPD reader script | MATLAB and ZIP | — | |||||||
| Clinical Data | ZIP and CSV | — |
Version 3: Updated 2022/01/26
A fractional shift in pixel positions was identified between the full dose and low dose Siemens’ patient cases included in Version 1 and 2. In the DICOM tag called “image position patient”, the full dose positions are written with seven digits after the decimal while the low dose positions have only two digits after the decimal and, in some cases, there is an offset close to 0.5 mm. This occurred due to having the full dose (original) data reconstructed on the scanner at the time of the patient exam and the low dose data reconstructed at a later time (after noise was inserted in the projection data) using Siemens’ off-line reconstruction tools. Note that this shift is a consequence of a difference in the reconstruction tools used. The projection data are not affected. The shift primarily impacted those using the full dose and low dose images to train machine learning algorithms.
The Siemens image data have been reprocessed so that the pixel shift issue has been addressed. The DICOM tag “image patient position” is now identical for the full dose and low dose images. The previous version of the data has been replaced with the new data (Version 3); click here to download a spreadsheet that maps the old UIDs to the new UIDs. You can access the updated series by downloading this manifest.
| Title | Data Type | Format | Access Points | License | Metadata | ||||
|---|---|---|---|---|---|---|---|---|---|
| Images | CT | DICOM | Download requires NBIA Data Retriever |
151 | 301 | 601 | 10,115,379 | — | |
| Images Phantom Object Only | CT | DICOM | Download requires NBIA Data Retriever |
1 | 1 | 1 | 18,032 | — | |
| DICOM-CT-PD User Manual Version 3 | — | ||||||||
| Matlab DICOM-CTPD data dictionary | TXT | — | |||||||
| Matlab DICOM-CTPD reader script | MATLAB and ZIP | — | |||||||
| Clinical Data | ZIP and CSV | — |
Version 2: Updated 2020/08/11
Important information about the GE patient cases.
An incorrect value was found in the RescaleIntercept DICOM tag (0028,1052) in the GE data. Additionally, we have been asked by users to determine and add PhotonStatistics values to DICOM tag (7033,1065). Hence, access to the GE data (149 cases) has been temporarily suspended while we address these issues.
- If you have already downloaded the GE data, you will want to download the updated data when it is again available.
- The GE data are readily recognized, as they have only 3 series per patient case (Siemens data have 4).
- The GE data can also be identified using the DICOM tag (0008,0070).
- If you have already started a project with the GE data, please contact the Mayo team to discuss the issues in greater detail to discern if they impact your work. Please send your inquiry to CTCIC@mayo.edu and the team will follow up with you.
| Title | Data Type | Format | Access Points | License | Metadata | ||||
|---|---|---|---|---|---|---|---|---|---|
| Images | DICOM | — | |||||||
| Images Phantom Object Only | CT | DICOM | Download requires NBIA Data Retriever |
1 | 1 | 1 | 18,032 | — | |
| DICOM-CT-PD User Manual Version 3 | — | ||||||||
| Matlab DICOM-CTPD data dictionary | TXT | — | |||||||
| Matlab DICOM-CTPD reader script | MATLAB and ZIP | — | |||||||
| Clinical Data | ZIP and CSV | — |
Version 1: Updated 2020/04/22
| Title | Data Type | Format | Access Points | License | Metadata | ||||
|---|---|---|---|---|---|---|---|---|---|
| Images | DICOM | Download requires NBIA Data Retriever |
— | ||||||
| Images Phantom Object Only | CT | DICOM | Download requires NBIA Data Retriever |
1 | 1 | 1 | 18,032 | — | |
| DICOM-CT-PD User Manual Version 3 | — | ||||||||
| Matlab DICOM-CTPD data dictionary | TXT | — | |||||||
| Matlab DICOM-CTPD reader script | MATLAB and ZIP | — | |||||||
| Clinical Data | ZIP and CSV | — |
